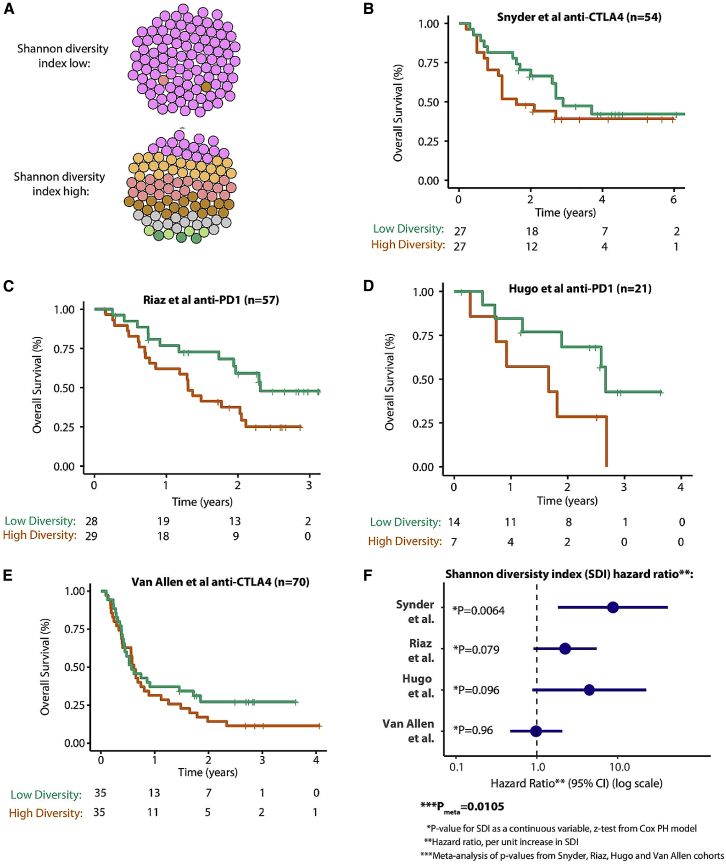
Figure 7.
Shannon Diversity Index (SDI) Analysis in Immune Checkpoint Inhibitor Datasets
(A) The cartoon illustrates two examples of the SDI, top low SDI (the tumor is predominantly composed of one major clone) and bottom high SDI (the tumor is composed of multiple clones with higher evenness between clones). SDI is measured using individual tumor subclones (from Pyclone clustering) as types and the somatic mutations as entities so that a tumor with a low SDI would have nearly all mutations concentrated in just one clone, and, in contrast, a tumor with a high SDI would have a higher number of clones, with mutations spread evenly or diversely across each clone.
(B) The SDI analysis applied to the Snyder et al. (2014) anti-CTLA4 dataset. Overall survival Kaplan-Meier plots are shown for with patients with a high SDI in red (SDI above median value in cohort) and a low SDI in green. The number of patients at risk by time point is shown in the table below.
(C–E) The same data format as in (B) for the Riaz et al. (2017) anti-PD-1 dataset (C), Hugo et al. (2016) anti-PD-1 dataset (D), and Van Allen et al. (2015) anti-CTLA4 dataset (D), respectively.
(F) Forest plot showing the HR for the SDI in each dataset, with the HR value corresponding to the survival risk per unit increase (i.e., each +1 increment) in the SDI. For significance analysis, SDI is tested as a continuous variable (to show a continuous association across the full range of data) using a Cox proportional hazard model (other clinical predictors, e.g., stage, are not included).