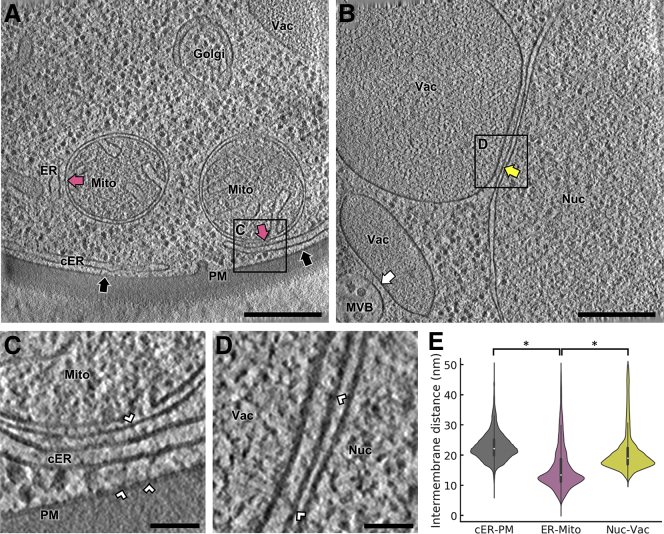
Figure 1.
Cryo-ET Imaging of MCS in WT S. cerevisiae
(A) 1.4 nm-thick tomographic slice showing cER-PM MCS (black arrows) and ER-mitochondria MCS (purple arrows). The boxed area is magnified in (C). ER: endoplasmic reticulum; cER: cortical ER; Golgi: Golgi apparatus; Mito: mitochondrion; PM: plasma membrane; Vac: vacuole.
(B) 1.4 nm-thick tomographic slice showing a nucleus-vacuole junction (yellow arrow) and a multivesicular body-vacuole MCS (white arrow). The boxed area is magnified in (D). MVB: multivesicular body; Nuc: nucleus.
(C) Magnification of the area boxed in (A). White arrowheads: intermembrane tethers.
(D) Magnification of the area boxed in (B).
(E) Violin plots showing the distribution of intermembrane distances of cER-PM, ER-mitochondrion and nucleus-vacuole MCS. The plots show the complete distribution of values including all MCS analyzed. A white dot represents the median, a black slab the interquartile range, and a black line 1.5x the interquartile range. ∗ indicates p < 0.05 by unpaired t test. N = 6 (cER-PM), 5 (ER-mitochondria) and 5 (nucleus-vacuole) MCS in WT cells. Scale bars: 300 nm (A, B), 50 nm (C, D). See also Figure S2; Table S1.