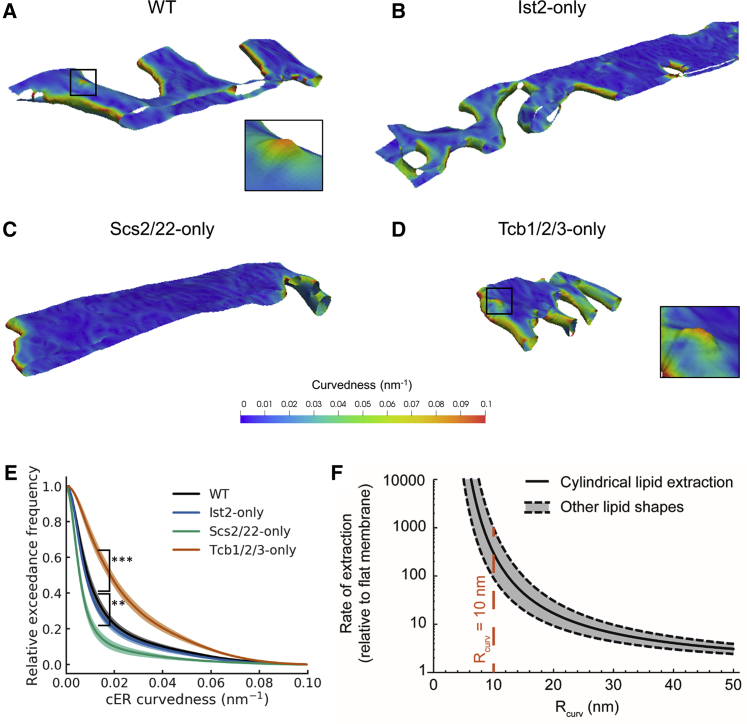
Figure 3.
Quantification of cER Curvature
(A–D) 3D visualizations of cER curvedness in the indicated strains. Insets in (A) and (D) show cER peaks. (A) WT cell, (B) Ist2-only cell, (C) Scs2/22-only cell, (D) Tcb1/2/3-only cell. (E) Quantification of cER curvedness, shown as an exceedance plot. The shaded lines represent the average across all MCS ± SE for each bin (1 nm-1.). ∗∗ and ∗∗∗ indicate, respectively, p < 0.01 and p < 0.001 by unpaired t test. N = 6 (WT), 7 (WT HS), 5 (Ist2-only), 5 (Scs2/22-only), and 9 (Tcb1/2/3-only) cER-PM MCS. (F) Enhancement of the rate of lipid extraction by membrane curvature according to a theoretical model. The plot shows the rate of extraction computed for a standard cylindrical lipid (black curve) as well as for lipids of other shapes, such as conical or inverted conical lipids (gray-shaded area between the dashed, black curves). The value of the radius of curvature of the experimentally observed cER peaks is denoted by the dashed red line. is equivalent to the curvedness for κ1=κ2. See also Figures S3 and S4; Table S1.