Figure 1.
Cell Rear Dynamics in Matrix-Directed Cells
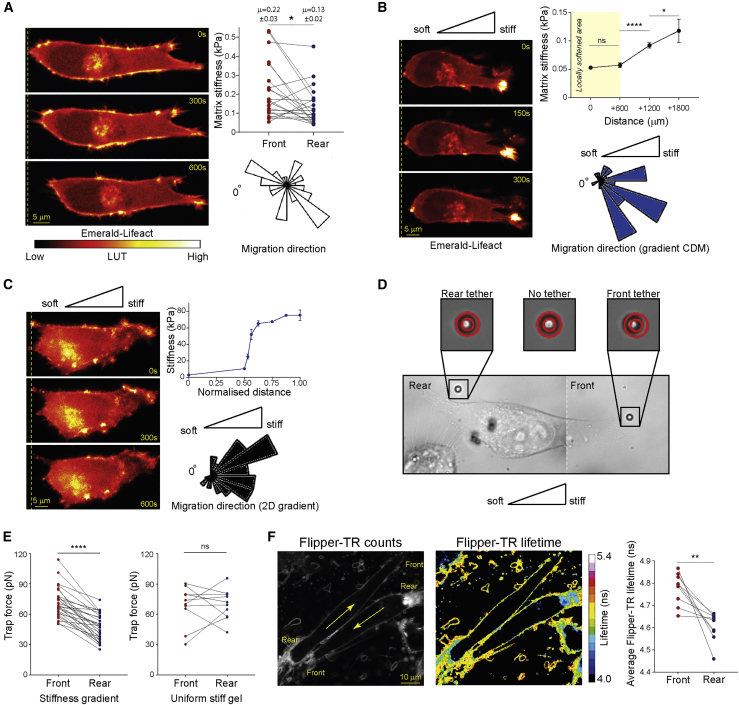
(A) A2780 cells in 3D CDM, confocal sections are shown 300 s apart. Dashed yellow line indicates the rearmost part of the cell in frame 1; Top right: Matrix rigidity immediately in front of and behind cells assessed by AFM, N = 20 cells (3 repeats); bottom right: orientation of migration shown by rose plot (N>35 cells, 3 repeats).
(B) Cells imaged as in (A) in gradient softened CDM; top right: rigidity across gradient softened CDM assessed by AFM before seeding cells (N=3; bars = SEM); bottom right: orientation of migration (locally softened matrix at 0°) shown by rose plot (N>35 cells, 3 repeats).
(C) A2780 cells on 2D FN-coated durotactic gradients imaged as in (A); top right: rigidity of 2D durotactic gradients assessed by AFM (N=3; bars = SEM); bottom right: orientation of migration with respect to the gradient shown by rose plot (N=58 cells, 3 repeats).
(D) Representative images from optical trap membrane tether experiments; white dotted line indicates separation of images.
(E) Paired trap force measurements at the front and rear of cells on durotactic gradients (left; N=29 cells, 3 repeats); or on uniform stiff gels (right; N=10, 3 repeats).
(F) Left, center: Cells migrating in CDM stained with Flipper-TR membrane tension probe; photon counts per pixel (left) and fluorescent lifetime imaging (FLIM) lifetime per pixel with a 16-color LUT (center); right: pairwise average Flipper-TR lifetime in manually identified membrane regions at the front and rear of cells (N=9 cells, 3 repeats; lower lifetime corresponds to lower membrane tension). Yellow arrows indicate direction of movement. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗∗p < 0.0001; ns, not significant. See also Figure S1 and Videos S1 and S2.