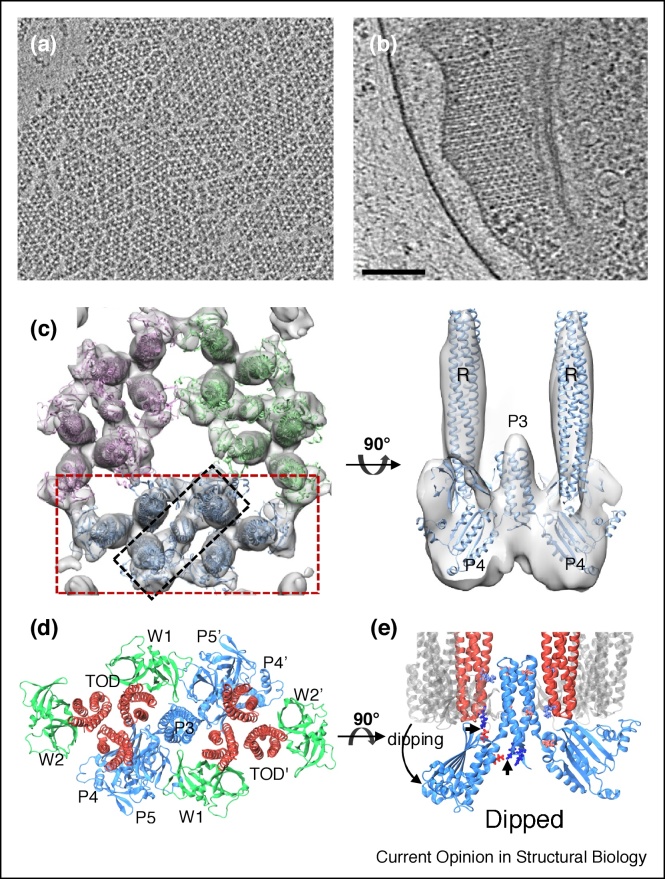
Figure 2.
CryoSTAC of a reconstituted bacterial chemotaxis signaling array.
(a) and (b) Tomographic slices of the in vitro reconstituted array of the chemotaxis core signaling complex (a) and of a native array in an Escherichia coli cell (b). Scale bar, 100 nm. (c) The density map of a threefold assembly unit by subtomogram averaging, with a core signaling complex boxed in red and a rotated view shown on the right corresponding to the black box. The receptor, P3 and P4 domains of Che A are labeled as ‘R’, ‘P3’ and ‘P4’, respectively. (d) Pseudo-atomic model of the core signaling complex, consisting of six chemoreceptor dimers (red, labeled ‘TOD’), one CheA dimer (blue, labeled ‘P3–P5’), and four CheW monomers (green, labeled ‘W1’ and ‘W2’). (e) CheA conformational dynamics with a dipping motion, determined by large scale MD simulation. Arrows point to the interacting amino acids in the dipped state [44••].