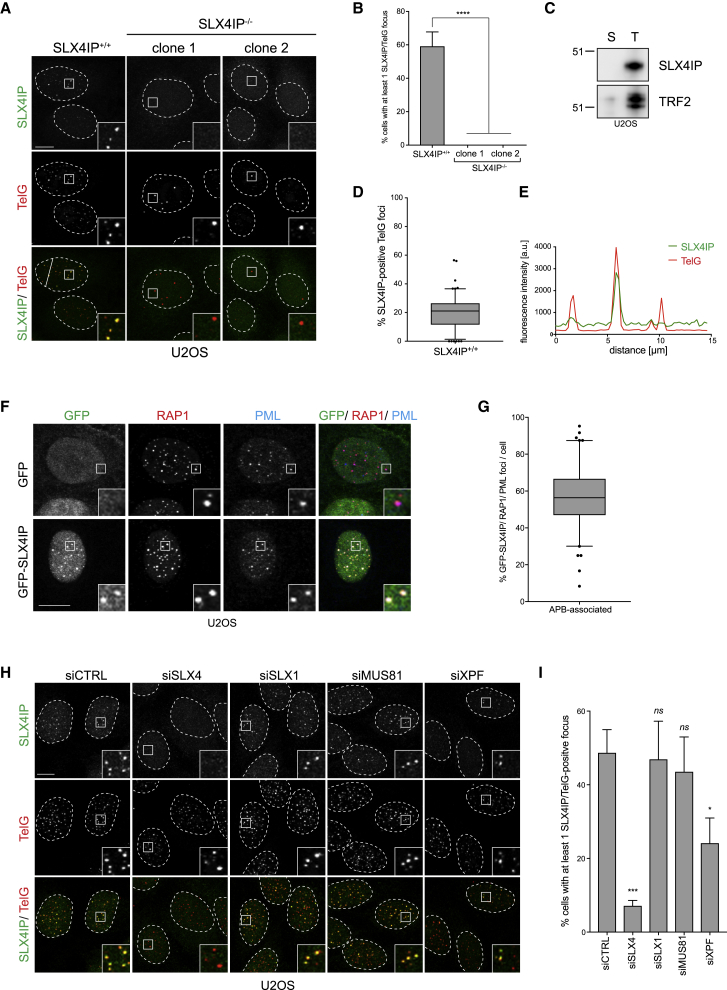
Figure 1.
SLX4IP Localizes at Telomeres in an SLX4-Dependent Manner
(A) U2OS cells were fixed and processed for SLX4IP immunofluorescence followed by telomeric PNA (TelG) FISH. Scale bar represents 10 μm. Line across the nucleus in SLX4IP+/+ indicates line profile measured in (D). Dashed lines indicate nucleus outlines (as determined using DAPI staining; not shown). Insets represent 3× magnifications of the indicated fields.
(B) Quantification of (A). At least 100 cells per condition were counted. Data are represented as mean ± SD; n = 3; ∗∗∗∗p < 0.00001, Student’s t test.
(C) Chromatin was isolated from whole-cell U2OS extracts with either a scrambled control (S) or a telomere-specific (T) 2′F-RNA probe. The chromatin was separated using SDS-PAGE and analyzed using SLX4IP immunoblotting. TRF2 was used as a telomeric chromatin control. Numbers denote molecular weight (kDa).
(D) Quantification of (A). At least 70 cells per experiment were counted. Data are represented as mean ± SD; n = 3.
(E) A random straight line was drawn across through a single Z section of the nucleus shown in SLX4IP+/+ in (A). The intensity of SLX4IP and TelG (telomeric PNA probe) was quantitated along the length of the line to generate a line profile.
(F) U2OS cells transfected with GFP or GFP-SLX4IP were fixed and processed for GFP, RAP1, and PML immunofluorescence. Scale bar represents 10 μm. Insets represent 3× magnifications of the indicated fields.
(G) Quantification of (F). At least 50 cells per condition were counted. Data are represented as mean ± SD; n = 3.
(H) U2OS cells were transfected with the indicated small interfering RNAs (siRNAs), fixed and processed for SLX4IP immunofluorescence followed by telomeric PNA (TelG) FISH. Scale bar represents 10 μm. Dashed lines indicate nucleus outlines (as determined using DAPI staining; not shown). Insets represent 3× magnifications of the indicated fields.
(I) Quantification of (E). At least 100 cells per condition were counted. Data are represented as mean ± SD; n = 3; ∗p < 0.01 and ∗∗∗p < 0.0001, Student’s t test.
See also Figures S1–S3.