Figure 7.
SLX4IP Is Lost in a Subset of ALT-Positive Osteosarcomas
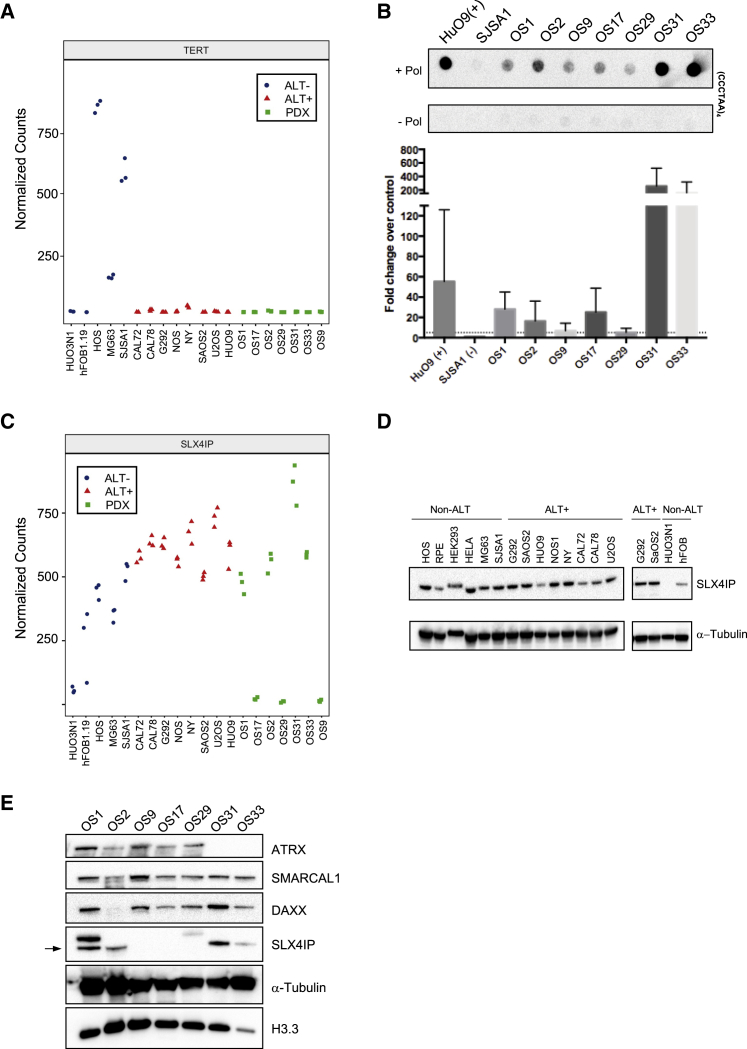
(A) Relative expression of hTERT from RNA sequencing preformed on a panel of osteosarcoma cell lines and patient-derived osteosarcoma xenografts. RNA sequencing was performed in triplicate, and each dot represents a separate experiment.
(B) Quantification of C-circle abundance in the osteosarcoma PDX samples. DNA was extracted from three separate tissue sections taken from each tumor. DNA extracted from HUO9 cells was used as a positive control, and DNA extracted from SJSA1 xenografts was used as a negative control. Data are represented as mean ± SD; n = 3. Dotted line represents 5-fold change in C-circle abundance.
(C) Relative expression of SLX4IP from RNA sequencing preformed on a panel of osteosarcoma cell lines and patient-derived osteosarcoma xenografts. RNA sequencing was performed in triplicate, and each dot represents a separate experiment.
(D) Whole-cell extracts of the indicated cell lines were analysed by SLX4IP immunoblotting. α-Tubulin was used as loading control.
(E) PDX tumor samples were analysed by SLX4IP, ATRX, SMARCAL1, DAXX and H3.3 immunoblotting. α-Tubulin was used as loading control. Arrow indicates SLX4IP band.
See also Figure S7.