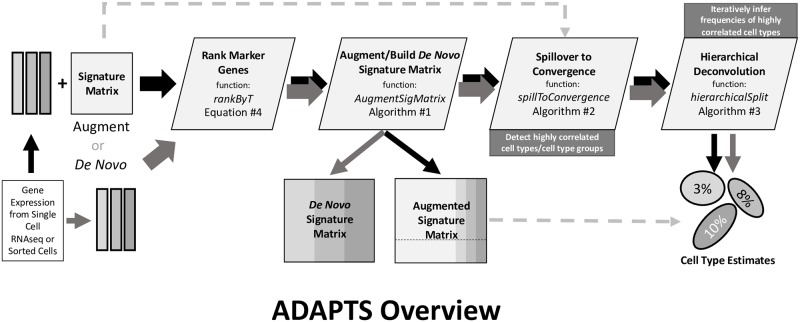
Fig 1. Overview of ADAPTS modules.
New gene expression data from cell types (e.g. from a tumor microenvironment) will be used to construct a new signature matrix de novo or by augmenting an existing signature matrix. First ADAPTS will rank marker genes for the cell types using the function rankByT described in Eq 4. Then ADAPTS adds marker genes in rank order using the function AugmentSigMatrix as described in Algorithm 1 resulting in a new signature matrix. This matrix may be tested for spillover between cell types using the function spillToConvergence described in Algorithm 2. Finally, ADAPTS separates cell types with heavy spillover using the hierarchical deconvolution function hierarchicalSplit described in Algorithm 3 to estimate the percentage of cell types present in bulk gene expression data.