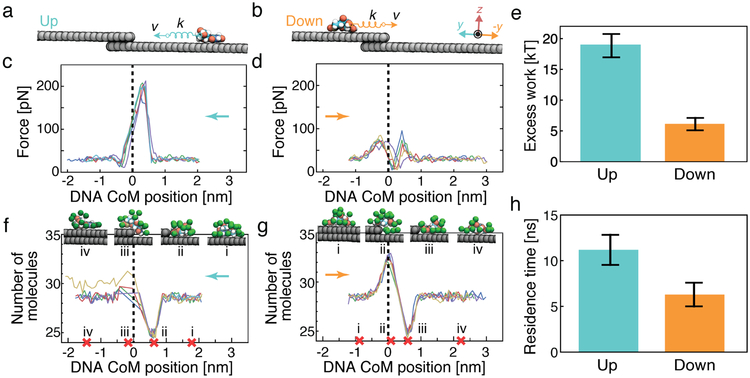
Figure 4:
Hydrophobic origin of the direction dependence of a defect crossing. (a,b) SMD simulation of a single nucleotide crossing a step defect. The CoM of a single nucleotide is pulled with a constant velocity of 0.1 nm ns−1 up or down a step defect. (c,d) Force applied to the DNA nucleotide by the SMD spring as a function of the nucleotide’s CoM coordinate in five independent SMD simulations. The instantaneous SMD forces were averaged according to the CoM coordinate in 0.1 nm bins. The step defect is located at the origin and is schematically indicated by a vertical dashed line. The arrow indicates the direction of the forced displacement. (e) Average excess work done when moving a nucleotide across a step defect. The work was determined by integration of the force–displacement curves. The excess work was computed as the total work done on the nucleotide by the SMD force in the [−1.0, +1.0] nm interval minus the work done when moving the nucleotide equivalent distance along a flat graphene surface. (f,g) Number of water molecules surrounding the nucleotide as function of nucleotide location. The number was computed using a 0.4 nm cutoff and averaged according to the CoM coordinate in 0.1 nm bins. The snapshots illustrate representative configurations of water molecules (green van der Waals spheres) corresponding to the nucleotide’s CoM coordinates marked by red crosses at the CoM axis. (h) Nucleotide residence time in the vicinity of the dehydration minimum defined as the [0.4, 0.8] nm interval along the CoM coordinate. Error bars represent standard deviations over five independent runs.