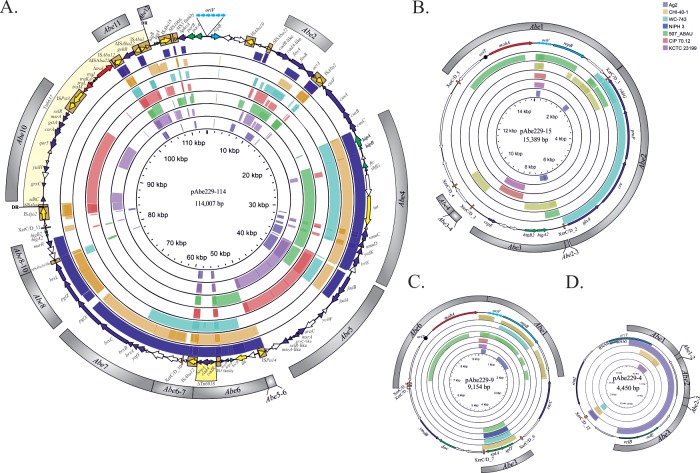
Fig 4. Identification of homologous sequences to HPC229 plasmids in other Acinetobacter genomes.
The colored arrows in the outer ring describe the location, identification, and direction of transcription of genes in the corresponding plasmids with described functions in databases. ORFs encoding for unknown functions are indicated by open arrows. In the subsequent rings, the regions of homology between the corresponding plasmid sequences and genome sequences of other A. bereziniae strains (indicated by arcs of different colors, see key near the upper right margin) as detected by BlastN searches are shown. The strains have been located from the highest (outer rings) to the lowest (inner rings) sequence coverage. The height of the colored arcs in each case is proportional to the percentage of nucleotide identity obtained in the BlastN search. BlastN hits with ≥70% nucleotide identity and a minimum alignment length of 1,000 bp (pAbe229-114) or 300 bp (pAbe229-15, pAbe229-9 and pAbe229-4) are detailed in Table 2. The darker lines in the arcs mark overlapping hits. The external Abe1-Abe12 grey regions indicate homologous sequences found in Acinetobacter non-bereziniae genomes (For details see Figs 1 and 2; and also Table 2).