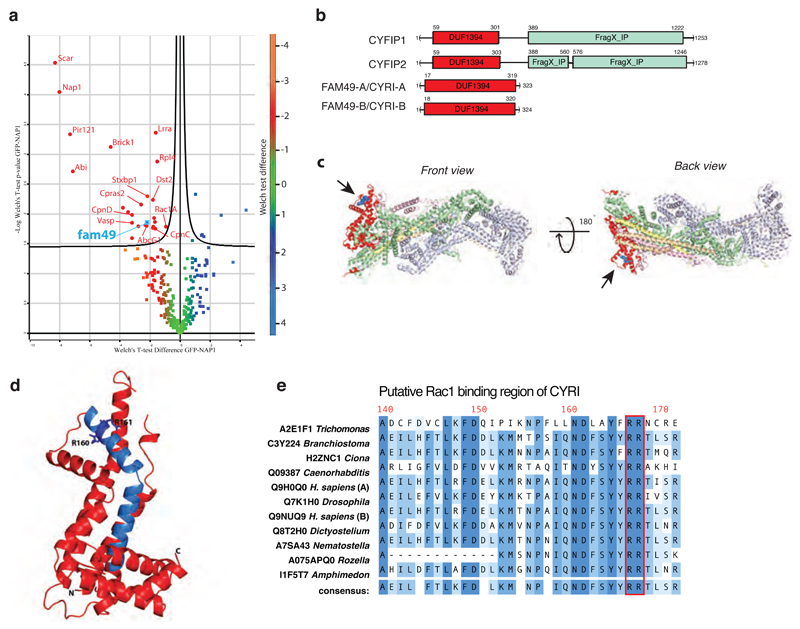
Figure 1. CYRI (Fam49) proteins show homology to CYFIP and contain a putative Rac1 interaction motif.
a – Volcano plot illustrating pooled results from four LC-MS/MS experiments showing comparison of formaldehyde crosslinked proteins co-immunoprecipitating with GFP or GFP-NAP1 in Dictyostelium napA knockout cells. Color-coding based on Welch test difference. Curved line is 5% false discovery rate. Identified interactors are labeled with gene symbols and presented in Table S2 with additional information.
b – Schematic of human CYFIP1/2 and CYRI-A/B showing amino acid numbers and domains. Common DUF1394 domain (Pfam PF07159) in red and CYFIP1/2 C-terminal cytoplasmic Fragile X Mental Retardation FMR1-interacting domain (FragX-IP, Pfam PF05994) in light green.
c - Two views of ribbon crystal structure of the Scar/WAVE complex (PDB 3P8C)2. NCKAP1 in lilac, CYFIP1 in light green and red, Scar/WAVE in peach, HSPC300 in yellow and ABI1 in orange. DUF1394 is red, with putative Rac1 interaction residues in blue and highlighted by arrows.
d – Phyre prediction of structure of the DUF1394 domain of CYRI-B. The putative Rac1-binding domain of CYRI is blue with Arg160 and Arg161 indicated as a stick representation.
e - Sequence alignment of the putative Rac1-binding domain of CYRI in different organisms. The CYFIP Lys189 and Arg190 equivalent residues are well conserved in CYRI (Arg160 and Arg161) and are highlighted in red.