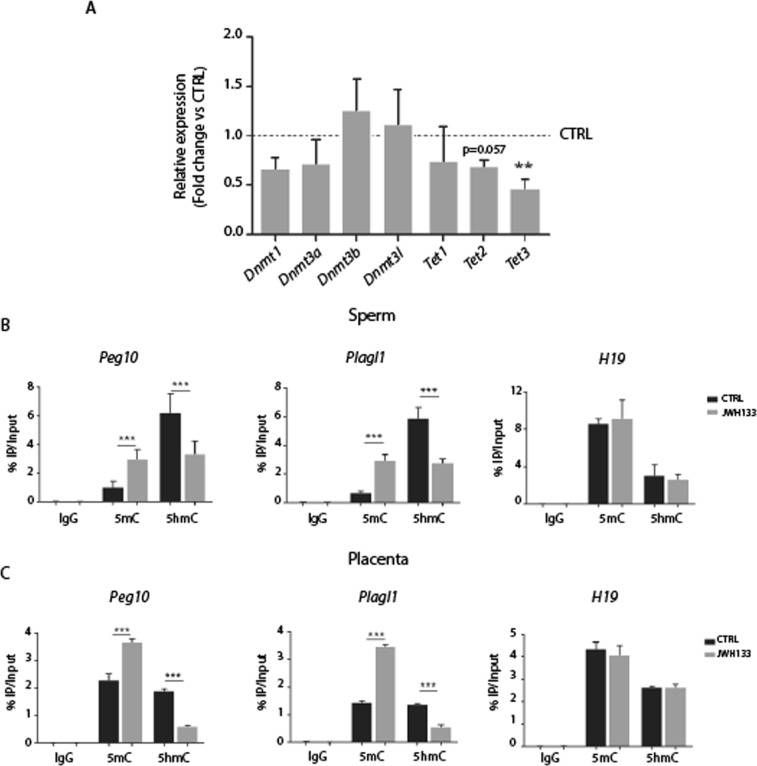
Figure 6.
Altered DNA methylation and hydroxymethylation levels in sperm from JWH-133 exposed males and in the placenta of their progeny. (A) Histogram reporting gene expression analysis of the DNA methyltransferases (Dnmts) and Tet hydroxylase performed by RT-qPCR on sperm RNA from JWH-133 exposed males (n = 6) with respect to controls (n = 6) that is set as 1 (Dnmt1: 1 ± 0.19; Dnmt3a: 1 ± 0.27; Dnmt3b: 1 ± 0.15; Dnmt3l: 1 ± 0.52; Tet1: 1 ± 0.41; Tet2: 1 ± 0.11; Tet3: 1 ± 0.09). b-Actin is used as reference gene. (B) MeDIP/hMeDIP analysis performed on pooled DNA obtained from sperm of six JWH-133 exposed males and six controls showing the enrichment of 5mC and 5hmC on imprinted genes. (C) MeDIP/hMeDIP analysis performed on pooled DNA obtained from placentas (n = 6) of three females crossed with JWH-133 exposed or control males. IgGs are used as controls and data obtained are shown as percentage of input DNA. Selected imprinted genes are Peg10 and Plagl1, paternally expressed and H19, maternally expressed. Results are reported as mean ± SD and statistical significance was obtained by Student’s t-Test; (**p < 0.01;***p < 0.001).