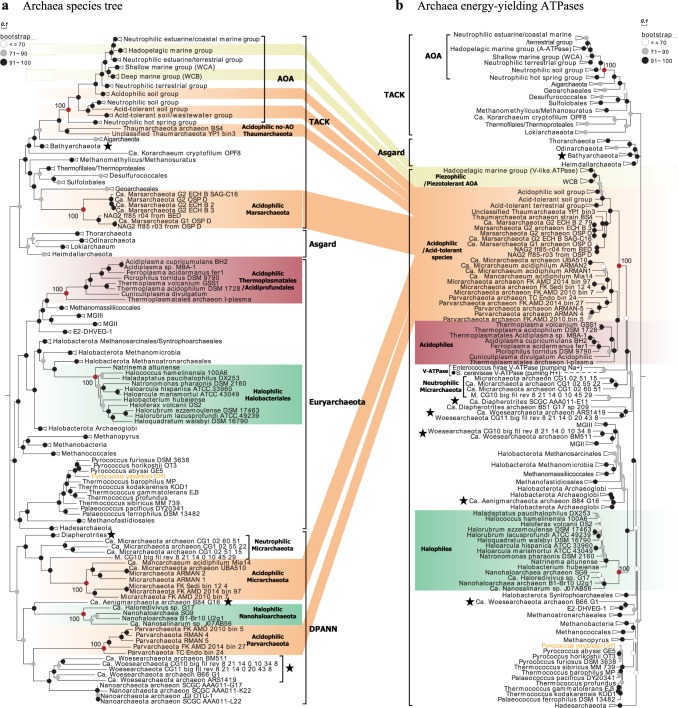
Fig. 3.
Comparative phylogeny of 36 conserved phylogenetic marker proteins and energy-yielding ATPases of Archaea domain. Only the genomes containing at least 30 of the selected 36 and 100 of the selected 122 conserved phylogenetic marker proteins as well as the whole atp operons were selected for comparative phylogenetic analysis (Figs. 1, S8, and S13). a Maximum likelihood species phylogenetic tree of archaea based on 36 conserved phylogenetic marker proteins [IQ-TREE [42] best-fit model LG+I+G4, 1000 ultrafast bootstraps]. Totally, 50 thaumarchaeotal genomes as well as 158 representative genomes of the phylum Euryarchaeota and superphyla TACK, Asgard, and DPANN were included. b Maximum likelihood tree of the ATPases based on the concatenation of the 7 major subunits (A, B, D, E, F, I, and K) present in all archaeal genomes [IQ-TREE [42] best-fit model, 1000 ultrafast bootstraps]. The V-ATPases from S. cerevisiae and E. hirae were also included in the phylogenetic analysis. The phylogenetic position of the S. cerevisiae V-ATPase (solid circles (●), dotted line) was inferred from the phylogeny of only subunits A and B, since its subunit compositions differed from archaeal ATPases. Some Archaeal branches whose positions in ATPase tree disagreed with their positions in the species tree are marked by stars