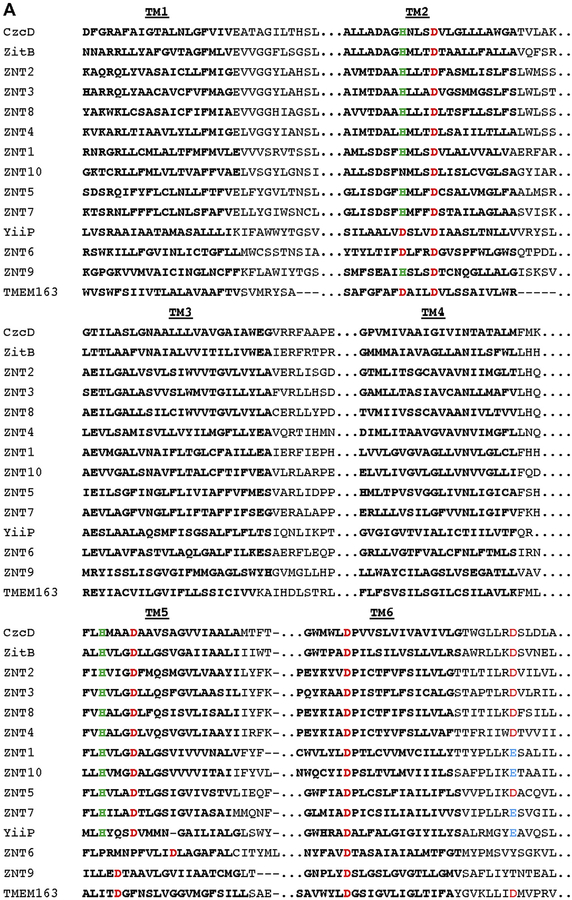
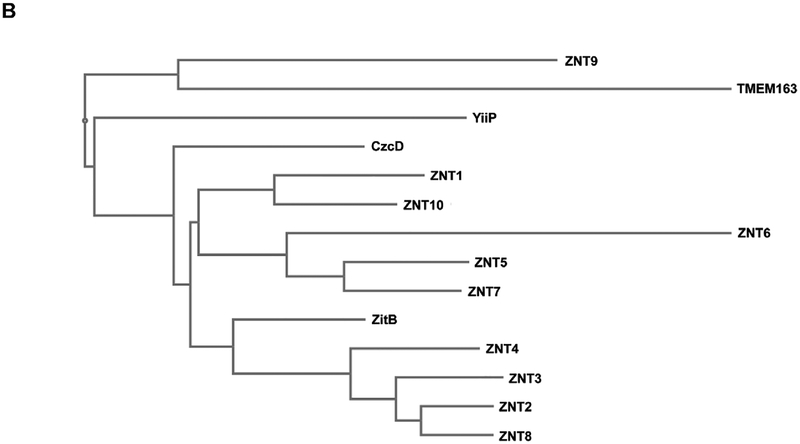
Fig. 2. Multiple sequence alignment of TMEM163 and select CDF family members.
A) Clustal alignment of TMEM163 and CDF amino acid sequences showing predicted transmembrane domains 1 through 6 (TM1-TM6; bold text). The alignment was done using the MAFFT website (https://mafft.cbrc.jp/alignment/server/) for the following full-length protein sequences obtained from NCBI: CzcD (YP_002515680.1), TMEM163 (NP_112185.1), YiiP (NP_462942), ZitB (WP_000951292), ZNT1 (NP_067017.2), ZNT2 (NP_001004434.1), ZNT3 (NP_003450.2), ZNT4 (NP_037441.2), ZNT5 (NP_075053.2), ZNT6 (NP_001180442.1), ZNT7 (NP_598003.2), ZNT8 (NP_776250.2), ZNT9 (NP_006336.3), and ZNT10 (NP_061183.2). For simplicity, only TM domains (bold text) and consensus or conserved amino acid residues predicted to bind zinc ions are shown. Histidine (green), Aspartic acid (red), and Glutamic acid (blue). B) Phylogram of the evolutionary relationship among specific CDF family members and TMEM163 protein. The phylogenetic tree diagram was generated using the Java-based Archaeopteryx visualization and analysis of phylogenetic trees via the MAFFT web interface.