Figure 1.
An Overall Increase in the Number of Differentially Expressed Transcripts across All DRGN Subpopulations with α-TOH Deficiency
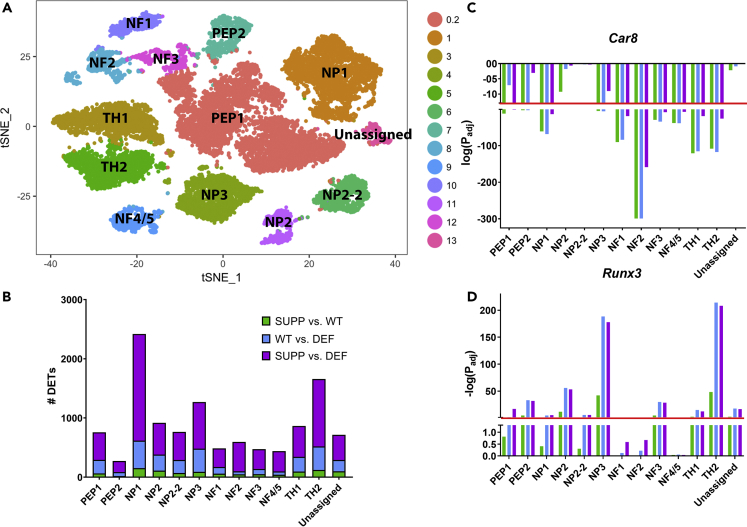
(A) A t-distributed stochastic neighbor embedding (t-SNE) plot of merged datasets of the six experimental mice to define neuronal subpopulations clusters. Thirteen clusters were identified based on previously reported gene expression profiles (Li et al., 2016, Usoskin et al., 2015). The “NP2-2” subgroup contained some genes representative of the NP2 cluster but was distinct from the NP2 cluster and has been previously characterized (Usoskin et al., 2015, Li et al., 2016).
(B) An increasing number of significantly (PFDR<0.05) differentially expressed transcripts (DETs) was identified in each DRGN subpopulation with increasing contrasts of vitE deficiency (i.e. SUPP vs. DEF > WT vs. DEF > SUPP vs. WT).
(C and D) (C) The two most commonly dysregulated transcripts across DRGN subpopulations were carbonic anhydrase 8 (Car8), which was significantly downregulated in 10/13 DRG clusters with vitE deficiency (SUPP vs. DEF), and (D) the lineage-specific transcription factor Runx3, which was significantly upregulated in 10/13 DRG clusters with vitE deficiency (SUPP vs. DEF). p values were adjusted by a false discovery rate of 0.05 and log-transformed. Significance was set at PFDR<0.05, corresponding to a –log Padjusted>1.3 (red line). NF = neurofilament, NP = non-peptidergic, PEP = peptidergic, TH = tyrosine hydroxylase, UNKNOWN = unknown cluster, n = 2 mice per group with ~3,600 cells/mouse profiled.