Abstract
Even though deficits in olfactory function affect a considerable part of the population, the neuronal basis of olfactory deficits remains scarcely investigated. To achieve a better understanding of how smell loss affects neural activation patterns and functional networks, we set out to investigate patients with olfactory dysfunction using functional magnetic resonance imaging (fMRI) and olfactory stimulation. We used patients’ scores on a standardized olfactory test as continuous measure of olfactory function. 48 patients (mean olfactory threshold discrimination identification (TDI) score = 16.33, SD = 6.4, range 6 ‐ 28.5) were investigated. Overall, patients showed piriform cortex activation during odor stimulation compared to pure sniffing. Group independent component analysis indicated that the recruitment of three networks during odor stimulation was correlated with olfactory function: a sensory processing network (including regions such as insula, thalamus and piriform cortex), a cerebellar network and an occipital network. Interestingly, recruitment of these networks during pure sniffing was related to olfactory function as well. Our results support previous findings that sniffing alone can activate olfactory regions. Extending this, we found that the severity of olfactory deficits is related to the extent to which neural networks are recruited both during olfactory stimulation and pure sniffing. This indicates that olfactory deficits are not only reflected in changes in specific olfactory areas but also in the recruitment of occipital and cerebellar networks. These findings pave the way for future investigations on whether characteristics of these networks might be of use for the prediction of disease prognosis or of treatment success.
Keywords: anosmia, fMRI, functional connectivity, hyposmia, neuronal plasticity, olfaction, olfactory disorders
1. INTRODUCTION
Anosmia, or the loss of the sense of smell, occurs in ∼5% up to 20% of the population, with an increase of this percentage with ageing (Boesveldt et al., 2017; Brämerson et al., 2004; Croy et al., 2014b; Rawal et al., 2016). Whereas smell ability is mostly assessed by using objective tests such as the Sniffin’ Sticks test (Hummel et al., 2007), structural and functional changes in the brain can also be used to explain and understand olfactory loss (for a review, see Reichert & Schöpf, 2017). Using functional magnetic resonance imaging (fMRI) to measure activity and functional connectivity during odor administration in patients with smell disorders will contribute to a fundamental understanding of how the olfactory system works and might lead to better predictions on prognosis and the effect of treatment options like olfactory training for patients suffering from smell loss.
Studies that investigate the effects of olfactory disorders on functional activity in the olfactory system during the administration of odors in general show decreased activation in olfactory areas of the brain (Vedaei et al., 2013). Changes in brain activity after smell loss have been investigated extensively in patients suffering from neurodegenerative disorders, like Alzheimer's disease (Cerf‐Ducastel & Murphy, 2003; Wang et al., 2010) and Parkinson's disease (Welge‐Lüssen et al., 2009), and in aging patients (Welge‐Lüssen, 2009). A drawback of these populations is that neuronal changes related to the disease but unrelated to smell loss might distort how smell loss and remaining function are reflected in the brain.
The olfactory system of the brain can be activated by the sensorimotor act of sniffing alone, even without the presence of an odor (Kollndorfer et al., 2014; Mainland & Sobel, 2006; Sobel et al., 1998a). Kollndorfer et al. (2015d) found more connections between brain regions responsible for processing olfactory stimuli in healthy controls than in anosmic patients during sniffing odorless air, although there was no difference in the spatial extent of the olfactory network between the groups. This indicates that sniffing and smelling are intertwined in healthy persons, but this connection seems affected in anosmic patients. Studies on repeated stimulation of the olfactory system, for example, by consciously smelling odors during olfactory training, show that this stimulation can lead to activation of the neuroplasticity capacities of the brain (Gilbert & Sigman, 2007; Kollndorfer et al., 2014; Sorokowska et al., 2015). Stimulation of the olfactory system can lead to improvement of olfactory function and concurrent changes in functional networks in patients who suffer from smell loss, indicating that smell loss is not always irreversible. For example, a study by Kollndorfer et al. (2015d) showed an increase in functional connectivity in response to chemosensory stimulation with a trigeminal compound after olfactory training in anosmic patients. This suggests that, even when patients are diagnosed with a smell disorder, functionality of the olfactory system in the brain might be maintained. However, it is not known how maintenance of the olfactory system is influenced by severity of olfactory loss. More knowledge on the neural networks within the olfactory system might lead to a better understanding of how and why olfactory training can lead to improvement of olfactory function (Patel, 2017) and to a better prediction of the effectiveness of olfactory training in diverse patient groups.
In this study, we set out to determine how decline in olfactory functioning affects neural activation patterns and networks in the olfactory system of the brain and how this is related to the severity of smell loss. While previous studies on brain activation in olfactory disorders have focused on comparing patients to healthy controls, in our study, we used patients’ scores on a standardized olfactory test as continuous measure of olfactory function, enabling us to assess the impact of the severity of the smell disorder on neural activation and networks. Moreover, as described above heterogeneous patient populations might be a confounding factor in previous investigations of the neuronal alterations after olfactory loss. Therefore, in this study, we only included patients who lost their sense of smell by causes that are not known to cause direct changes in the brain, like infection of the upper respiratory tract and sinonasal diseases (Temmel et al., 2002). Furthermore, previous studies indicated that group independent component analysis (ICA) can provide supplementary information in chemosensory stimulation studies in addition to model‐dependent analyses (Frasnelli et al., 2012; Kollndorfer et al., 2015b). Therefore, in addition to traditional general linear model analyses, we applied this approach to extract functionally connected networks. Thus, in this study, we investigated neural responses and functional networks during odor administration in a sample of anosmic and hyposmic patients.
2. METHODS
2.1. Patient Sample
A total of 124 patients suffering from olfactory dysfunction took part in the clinical care assessment offered by the Smell and Taste Centre at Hospital Gelderse Vallei (Ede, the Netherlands), in collaboration with the division of Human Nutrition of Wageningen University (Wageningen, the Netherlands). All patients visited the center between July 2015 and October 2016. Of this initial sample, 76 patients were not included in the present study for various reasons (MRI abnormalities: 14, head trauma: 24, chronic diseases including mental health problems and cardiovascular diseases: 8, incomplete MRI or behavioral data: 18, excessive movement artifacts: 3, congenital anosmics: 8, no olfactory deficit according to the olfactory testing: (1). Assessment of MRI abnormalities was based on patients’ clinical T2 scans and carried out by a radiologist. Patients exhibiting major neural alterations (such as tumors, severe white matter deviations, atrophies, or early signs of neurodegenerative diseases) were excluded. Patients suffering from posttraumatic smell loss were excluded as they might show neuronal changes unrelated to their olfactory deficits (Lötsch et al., 2016; Yousem et al., 1999). Congenital anosmics were not included as the low sample size did not allow treating them as a subgroup, and their neuronal processing might differ fundamentally from acquired anosmics. Thus, in this study, we analyzed data of 48 patients (29 anosmics and 19 hyposmics) suffering from olfactory loss. Patient sample characteristics are listed in Table 1. All patients gave permission for the use of their medical records for this study.
Table 1.
Detailed description of patient sample
| N | Age [mean (SD)] | Female/male | Disease duration [years] | Cause of olfactory dysfunction | |||||
|---|---|---|---|---|---|---|---|---|---|
| <2 | 2–10 | >10 | Postinfectious | Sinonasal | Idiopathic/other | ||||
| Hyposmic | 19 | 57.9(11.34) | 11/8 | 9 | 4 | 6 | 10 | 7 | 2 |
| Anosmic | 29 | 60.3(14.53) | 19/10 | 5 | 10 | 14 | 5 | 13 | 11 |
*“Other” includes ageing and medicine use.
2.2. Procedure
As part of the standard clinical care assessment, all patients participated in clinical established testing of olfactory function (Hummel et al., 1997; Kobal et al., 1996; see next section for details). The clinical assessment further comprised tests that were not included in the present analysis (such as assessment of gustatory function using “Taste Strips”; Mueller et al., 2003) and assessment of retronasal olfactory function as in Croy et al., 2014a). Moreover, an ENT physician performed a nasal endoscopy to examine nose and mouth of the patients and conducted a medical history review to determine possible cause and duration of the disorder. All included patients took part in structural and functional MRI measurements (Section 2.2.2). The use of clinically collected data for research purposes was approved by the local ethical committee (Review committee for scientific research of Hospital Gelderse Vallei, Ede, the Netherlands; BC/1703‐143). All patients provided written informed consent.
2.2.1. Assessment of olfactory function
Olfactory function was assessed according to the procedure described in Hummel et al. (1997). Patients were presented with pen‐like odor sticks (“Sniffin’ Sticks,” Burghart Instruments, Wedel, Germany) in three tasks, assessing odor detection threshold, odor discrimination, and odor identification ability. In the odor threshold task, patients had to determine repeatedly in a forced‐choice procedure which of three sticks contained a target odorant (n‐butanol). The odorant and two distractor sticks without an odor were presented in a staircase up and down procedure. Out of 7 reversals, the last 4 turning points were averaged to obtain the threshold score. In the discrimination task, 16 triplets of odorants were presented (two containing the same odorant, while one stick contained an aberrant odorant). Patients were instructed to point out the odd odorant in a forced‐choice procedure. During the threshold and discrimination task, patients were blindfolded. The odor identification task consisted of 16 odors. Patients had to select the right label for each odor from a list of four descriptors provided. Odor identification was assessed for each nostril separately. For the present analysis, the average score of both nostrils was used. Threshold scores range from 1 to 16, while the scores for the discrimination and identification tasks range from 0 to 16. The three subscores were summed up to obtain the total Threshold‐Discrimination‐Identification score (TDI score). Based on clinical definitions (Hummel et al., 2007), we distinguished anosmia (TDI score ≤ 16) and hyposmia (16 < TDI score < 30.3).
2.2.2. MRI Data Acquisition
All scans were acquired on a 3 T Siemens Magnetom Verio MRI scanner (Siemens, Erlangen, Germany; software version VB19), using a 32‐channel head coil, at Hospital Gelderse Vallei, Ede, the Netherlands. For all scans, GRAPPA factor 2 was used. A 2D echo‐planar imaging (EPI) sequence was used for collecting the functional data with 847 scans and 45 axial slices (slice thickness = 3 mm, matrix size of 64 × 64, TE/TR of 25/2,240 ms, FoV 192 × 192 mm2, 90° flip angle). The stack was tilted at an angle of 30° to the anterior–posterior commissure line for all patients. A sagittal T1‐weighted 2D isotropic MP‐RAGE scan (192 slices, TE/TR of 2.26/1,900 ms, slice thickness = 1 mm, FoV = 256 × 256 mm2, 9° flip angle) was acquired for anatomical reference.
Olfactory stimulation during the functional scan was performed using an 8‐channel computer‐controlled olfactometer (Burghart, Wedel, Germany). Odors were administered birhinally to the patient through 2 nose pieces that were placed in the nostrils of the patient. Two high caloric, pleasant food odors, equivalent in intensity and used in previous behavioral and fMRI studies (Griffioen‐Roose et al., 2014; Zoon et al., 2016), were used: a sweet odor, chocolate (IFF, 10810180; 8.5% dissolved in propylene glycol), and a savory odor, beef (IFF, 10878095), 0.04% dissolved in demineralized water). These odors are considered familiar and ecologically relevant; however, there were no a priori hypotheses for these specific odors within this experiment. Odor stimuli were embedded in a stream of odorless, humidified air (80%, air flow 8 L/min, 36°C). Stimulus duration of the odor pulse was 2,000 ms. As control, blank trials were incorporated, during which visual cues were presented in equal length as the odor trials, while no odor was presented.
The fMRI paradigm consisted of two blocks separated by one minute rest. In total, 20 chocolate odor trials, 20 beef trials and 20 blank trials were presented. Additionally, 10 combined chocolate & picture and 10 beef & picture trials were presented. During these trials, patients were shown a picture of a chocolate muffin (chocolate & picture trials) or a steak (beef & picture trials) in addition to the odor. All trials were equally divided between the two blocks and were randomized within the blocks. All trials were preceded by a white fixation cross turning red. Patients were instructed to sniff through the nose when they saw the red fixation cross (duration 3,200 ms). To sustain patients' attention, 30 of the odor/blank trials were followed by the question “How intense did you perceive the odor?” with the anchors “not strong at all” and “very strong.” Eleven of the combined odor & picture trials were followed by the question “How well did the picture and the odor match?” with the anchors “not matching at all” and “very matching.” Patients responded to these questions by moving a cursor along a visual analogue scale (VAS, range 0–100) by button presses on a button box with the thumb of the right hand. The cursor always started at the center of the VAS. Trials were presented with varying inter‐stimulus interval (ISI, between 11 and 20 s). Presentation of the visual cues and pictures and triggering of the olfactometer was done with the use of E‐Prime 2.0 (Psychology Software Tools Inc). See Figure 1 for details on stimulus timing.
Figure 1.
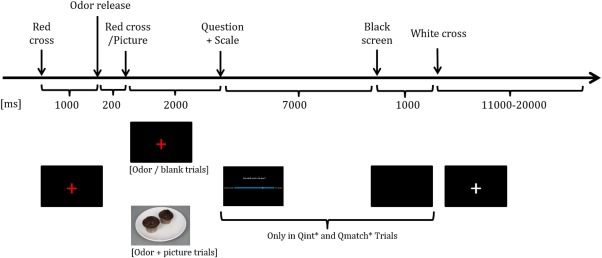
Details on stimulus timing during the olfactory paradigm. * QInt = intensity question (“How intense did you perceive the odor?”); QMatch = matching question (“How well did the picture and the odor match?”) [Color figure can be viewed at http://wileyonlinelibrary.com]
2.3. MRI data analysis
2.3.1. Processing of the fMRI data
Functional MRI data was preprocessed using SPM12 (http://www.fil.ion.ucl.ac.uk/spm/) implemented in Matlab R2011a, including slice‐time correction, motion correction, realignment, and spatial smoothing with a 6‐mm Gaussian kernel (full‐width at half‐maximum). Before second level analysis, the ArtRepair toolbox was used (Mazaika et al., 2009) to reduce the residual errors of more than 0.5 mm movement between scans which remained after realignment.
Our setup to investigate the preprocessed imaging data included two steps: We first investigated which activations were evoked by the odor stimuli compared to pure sniffing (during blank trials) by using a classical general linear model (GLM) analysis. In a second step, to investigate the functionally connected networks responsible for processing the sensory stimuli, we assessed group‐independent component analysis (ICA). Both analyses provide complimentary information when it comes to the investigation of functional task‐related imaging data. GLM reveals which activation patterns are evoked by the stimulus paradigm under the assumption of a regression model and the hemodynamic response function, whereas ICA is based on a blind sort separation algorithm without the use of an a priori paradigm, that separates functionally connected networks solely based on their temporal patterns. Please see other chemosensory research for reference (Frasnelli et al., 2012; Kollndorfer et al., 2015b, 2015c; Reichert et al., 2017).
2.3.2. GLM analysis
For subject‐level analysis, the following conditions were modeled: chocolate odor, beef odor, blanks, chocolate & picture, beef & picture trials and questions (intensity/matching). Six motion regressors were included as regressors of no interest. Subsequently, for each patient parameters were estimated for the comparison (subsequently referred to as “contrast”) of odor (chocolate and beef) to the blank condition. The odor (chocolate and beef) > blank contrast images of the patients were entered into a group‐level one‐sample t test to assess activation in response to olfactory stimulation across the whole sample. For this contrast, only the pure odor trials (without pictures) were used. Significance was assessed at a whole‐brain FWE corrected threshold (p fwe < .05). In a subsequent multiple regression analysis, we assessed whether overall olfactory function (regressor: TDI scores) was related to the odor‐related activity (odor > blank contrast images). For this analysis, a small volume correction (SVC, sphere of 20 voxels around peak of activation) statistical thresholding approach was applied. Moreover, a region of interest analysis of piriform cortex activation was carried out for both subgroups (hyposmics and anosmics) on the odor > blank contrast images to assess whether residual activation of piriform cortex was present for both subgroups. A functional mask for the piriform cortex from a meta‐analysis (Seubert et al., 2013) was used for these subgroup analyses and significance was assessed at a FWE‐corrected threshold (p fwe < .05).
2.3.3. Group ICA
We conducted group independent component analysis (ICA) on the preprocessed fMRI data using the GIFT toolbox (Calhoun et al., 2001). The number of components was estimated as 44 (mean of estimated components across all patients) using the minimum description length (MDL) criterion included in the group ICA of fMRI (GIFT) toolbox. Statistical reliability of independent components was assessed using the ICASSO method that validates the independent component time‐series via clustering and visualization (Himberg et al., 2003). Using ICASSO, the component estimation was performed 20 times with varying initial conditions of the algorithm. In a 2‐step principal component analysis (PCA) reduction procedure, components were reduced from 91 (maximum estimated by the implemented MDL algorithm) to 44. For group ICA, the Infomax algorithm was used. Subsequently, the extracted components were inspected visually and 14 artifactual components (overlapping substantially with known motion, susceptibility, vascular, or ventricular artifacts) were excluded.
In the next step, to assess which network was most related to odor processing, the network time courses were submitted to a multiple regression with a regressor specifying all odor presentation onsets (including pure odor trials and odor & picture trials). This step was carried out for each patient and resulted in individual beta weights for the odor regressor. The component C37 (subsequently referred to as “sensory processing network”) showed the highest task‐relatedness and contained a number of regions associated with olfactory processing previously, and was thus examined further. To examine this network in more detail and to compare odor stimulation to pure sniffing, the time course of the sensory processing network C37 was subjected to a further multiple regression including the odor onsets (beef and chocolate combined in one regressor) and blank onsets as regressors of interest. Six motion regressors and the other events of the paradigm (odor & picture onsets, onsets of questions on matching/intensity) were additionally included in the regression model as regressors of no interest. The resulting beta weights of the odor and blank regressors (reflecting the extent to which the network's time course was related to these two regressors) were subsequently correlated with TDI scores to assess whether network recruitment during these trials was related to patients' olfactory function.
In the final step, in an explorative analysis, we examined whether any additional network besides C37 was associated with sensory processing and olfactory function. Thus, the time courses of each of the remaining 29 (non‐artefactual) components were subjected to the multiple regression model described above (model with odor onsets, blank onsets, odor + picture onsets, onsets of questions on matching/intensity). Task‐relatedness was defined as a significant beta‐weight in a one‐sample t test (p < .05 after FDR‐correction to correct for the number of components tested). 19 task‐related components emerged from this analysis. For these components, beta weights of odor and blank trial regressors were correlated with TDI scores and evaluated for significance (p < .05 after FDR‐correction to correct for the number of components tested).
The averaged spatial component maps of the components of interest were entered into a one‐sample t test, thresholded at p < .05, FWE‐corrected to determine the main brain regions comprised in the component maps. For visualization of fMRI analyses results in Figures 3, 4, 5, whole‐brain component maps were exported to the “Multi‐image analysis GUI” (MANGO, http://ric.uthscsa.edu/mango) and overlaid on a standard anatomical template in MNI space.
Figure 3.
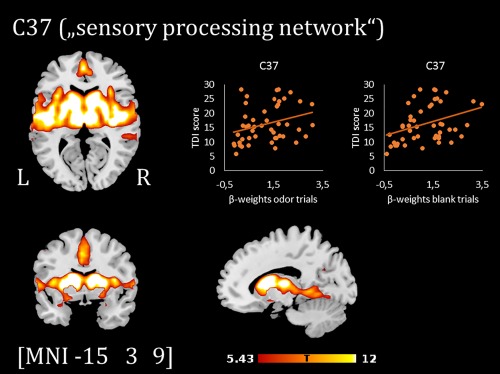
Component map of the sensory processing network (C37, thresholded at p < .05 FWE‐corrected) and scatterplots showing the positive correlation between TDI scores and β‐weights of the regressor odor/blank trials [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 4.
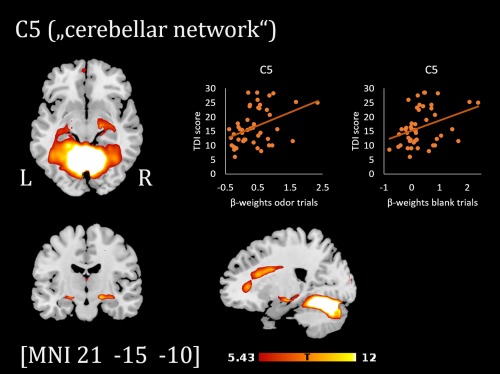
Component map of the cerebellar network (C5, thresholded at p < .05 FWE‐corrected) and scatterplots showing the relation between TDI scores and β‐weights of the regressor odor/blank trials [Color figure can be viewed at http://wileyonlinelibrary.com]
Figure 5.
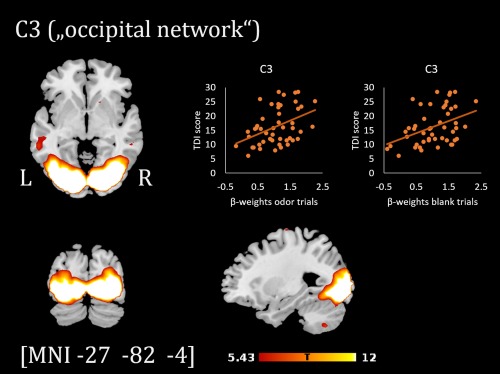
Component map of the occipital network (C3, thresholded at p < .05 FWE‐corrected) and scatterplots showing the relation between TDI scores and β‐weights of the regressor odor/blank trials [Color figure can be viewed at http://wileyonlinelibrary.com]
2.4. Intensity and matching ratings
Behavioral ratings were analyzed with IBM SPSS Statistics (version 24). Ratings were first averaged per patient. Average intensity ratings for odors versus blanks were compared using a paired T test (combining hyposmic and anosmic patients). To compare odor intensity ratings from anosmic patients to hyposmic patients, only questions for beef and chocolate odor were included (blanks were excluded), using an independent‐samples T test.
Pearson correlation was used to assess the relationship between Sniffin’ Sticks score (TDI) and averaged odor intensity ratings per patient.
Ratings of the matching questions were compared between hyposmic and anosmic patients using an independent‐samples T test.
3. RESULTS
3.1. Odor intensity and matching of odors and pictures
The group as a whole (n = 48) rated odors (mean 24.9 ± 25.6) as more intense than blanks (11.2 ± 14.8; p < .001). Hyposmic patients rated the odors with higher intensity than anosmic patients (41.3 ± 23.8 vs 14.2 ± 21.0; p < .001).
Olfactory function scores (TDI) were significantly positively correlated to odor intensity ratings (r = .45, p = .001).
Hyposmic patients rated the match between the odor and (congruent) picture as higher (55.3 ± 20.4) than the anosmic patients (33.7 ± 20.1; p < .001).
3.2. Olfactory activation in hyposmics and anosmics (GLM)
The results of the whole‐brain one‐sample t test showed an increased activation in the piriform cortex (Figure 2 and Table 2) for odor trials compared to the pure sniffing trials (blank trials), approaching significance at a whole‐brain FWE‐corrected peak threshold level (p FWE = .055). No significant relation emerged from the multiple regression analysis with TDI scores (p FWE > .1). ROI‐based one‐sample t tests indicated that there was piriform cortex activation for odors compared to blank trials in both subgroups (hyposmics and anosmics; see Table 2).
Figure 2.
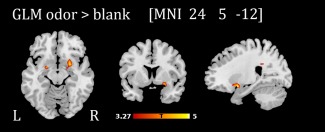
Activation pattern in the piriform cortex during olfactory stimulation compared to blank trials (results of one‐sample t test on contrast images odor > blank trials for whole sample, n = 48). For illustration purposes, activations are shown at p unc. < .001 [Color figure can be viewed at http://wileyonlinelibrary.com]
Table 2.
Significant clusters of activation for hyposmics and anosmics during the olfactory paradigm (odor > blank trials)
| Brain region (peak/nearest grey matter) | Side | Peak MNI coordinates | Peak T value | Cluster size | p FWE | ||
|---|---|---|---|---|---|---|---|
| x | y | z | |||||
| Whole‐brain, whole sample odor > blank | |||||||
| Piriform cortex | R | 24 | 2 | −16 | 5.35 | 32 | .055a |
| ROI analysisb, hyposmics odor > blank | |||||||
| Piriform cortex | R | 27 | 2 | −16 | 6.03 | 2 | <.001 |
| L | −21 | −4 | −10 | 4.80 | 3 | .006 | |
| R | 18 | −1 | −10 | 4.25 | 1 | .019 | |
| ROI analysisb, anosmics odor > blank | |||||||
| Piriform cortex | L | −21 | 2 | −16 | 4.48 | 4 | .004 |
| R | 21 | 8 | −16 | 3.99 | 8 | .013 | |
Note. Abbreviations: L = left; MNI = Montreal Neurological Institute; R = right.
Results of one‐sample t tests.
Results corresponding to p < .001 uncorrected significance level with p < .055 peak activation.
ROI‐analysis = region of interest analysis using a mask of the piriform cortex from [Seubert et al., 2013]. For ROI‐analyses, only activations significant at a height‐level threshold of p < .05 with family‐wise error (FWE) correction are displayed. For whole‐brain comparison, activations significant at p < .001 uncorrected are reported.
3.3. Recruitment of networks during odor administration (group ICA)
Figure 3 shows the spatial extent of the sensory processing network C37, comprising amongst others insula, thalamus, piriform cortex, cingulate gyrus, and putamen (see Table 3 for the top brain regions of the component map). Recruitment of this network during odor trials was positively correlated with total TDI scores (r(46) = .30, p = .039), as well as recruitment during blank trials (r(46) = .37, p = .011).
Table 3.
Main brain regions included in the three component maps
| Component | Standardized region name | AAL region | No of voxels |
|---|---|---|---|
| C37 (“sensory processing network”) | Median cingulate and paracingulate gyri (R) | Cingulum_Mid_R | 452 |
| Median cingulate and paracingulate gyri (L) | Cingulum_Mid_L | 421 | |
| Superior frontal gyrus, medial (L) | Frontal_Sup_Medial_L | 307 | |
| Superior temporal gyrus (L) | Temporal_Sup_L | 283 | |
| Insula (L) | Insula_L | 270 | |
| Insula (R) | Insula_R | 241 | |
| Anterior cingulate and paracingulate gyri (L) | Cingulum_Ant_L | 237 | |
| Thalamus (L) | Thalamus_L | 237 | |
| Middle temporal gyrus (L) | Temporal_Mid_L | 225 | |
| Thalamus (R) | Thalamus_R | 220 | |
| Lenticular nucleus, putamen (L) | Putamen_L | 220 | |
| Supplementary motor area (L) | Supp_Motor_Area_L | 217 | |
| Superior temporal gyrus (R) | Temporal_Sup_R | 209 | |
| Anterior cingulate and paracingulate gyri (R) | Cingulum_Ant_R | 194 | |
| Lenticular nucleus, putamen (R) | Putamen_R | 190 | |
| Rolandic operculum (L) | Rolandic_Oper_L | 176 | |
| Lingual gyrus (L) | Lingual_L | 166 | |
| Supplementary motor area (R) | Supp_Motor_Area_R | 156 | |
| Lingual gyrus (R) | Lingual_R | 150 | |
| Rolandic operculum (R) | Rolandic_Oper_R | 149 | |
| Caudate nucleus (L) | Caudate_L | 139 | |
| Caudate nucleus (R) | Caudate_R | 137 | |
| Middle temporal gyrus (L) | Temporal_Mid_R | 130 | |
| Superior frontal gyrus, medial (R) | Frontal_Sup_Medial_R | 113 | |
| C3 (“occipital network”) | |||
| Middle occipital gyrus (L) | Occipital_Mid_L | 526 | |
| Middle temporal gyrus (L) | Temporal_Mid_L | 319 | |
| Lingual gyrus (L) | Lingual_L | 261 | |
| Lingual gyrus (R) | Lingual_R | 258 | |
| Middle occipital gyrus (R) | Occipital_Mid_R | 252 | |
| Inferior occipital gyrus (L) | Occipital_Inf_L | 207 | |
| Calcarine fissure and surrounding cortex (L) | Calcarine_L | 202 | |
| Calcarine fissure and surrounding cortex (R) | Calcarine_R | 198 | |
| Fusiform gyrus (L) | Fusiform_L | 187 | |
| Fusiform gyrus (R) | Fusiform_R | 156 | |
| Precentral gyrus (L) | Precentral_L | 146 | |
| Superior occipital gyrus (R) | Occipital_Sup_R | 114 | |
| Superior occipital gyrus (L) | Occipital_Sup_L | 112 | |
| C5 (“cerebellar network”) | |||
| Hemispheric lobule VI (L) | Cerebellum_6_L | 378 | |
| Hemispheric lobule VI (R) | Cerebellum_6_R | 352 | |
| Crus I (L) | Cerebellum_Crus1_L | 313 | |
| Fusiform gyrus (L) | Fusiform_L | 311 | |
| Crus I (R) | Cerebellum_Crus1_R | 280 | |
| Fusiform gyrus (R) | Fusiform_R | 241 | |
| Hemispheric lobule IV/V (L) | Cerebellum_4_5_L | 222 | |
| Lingual gyrus (L) | Lingual_L | 222 | |
| Lingual gyrus (R) | Lingual_R | 165 | |
| Vermic lobule IV/V | Vermis_4_5 | 142 | |
| Hemispheric lobule IV/V (R) | Cerebellum_4_5_R | 133 |
Note. Abbreviations: L = left, R = right.
Brain regions classified by AAL (automatic anatomic labeling) atlas (http://www.gin.cnrs.fr/AAL?lang = en; Tzourio‐Mazoyer et al., 2002). Labeling was conducted on binary masks of the thresholded component maps (at <ip>FWE < .05, <ik> = 100). Please note that for this reason, no voxel intensity information is provided.
The beta weights for odor trials of two further components showed a significant correlation with TDI scores at p FDR < .05): C3 (subsequently termed “occipital network”): r(46) = .42, p = .003 and C5 (subsequently termed “cerebellar network”): r(46) = .41, p = .004. For both components, beta weights for blank trials were positively correlated with TDI scores as well (C3: r(46) = .41, p = .004 and C5: r(46) = .35, p = .014). The spatial component maps of these two networks are shown in Figures 4 and 5 (see Table 3 for the top brain regions comprised in the component maps).
4. DISCUSSION
In this study, we set out to determine for the first time how the severity of olfactory loss is reflected in functional brain activity and brain networks. To this end, we investigated neuronal activation in response to olfactory stimulation and pure sniffing in patients suffering from varying degrees of olfactory deficits. We applied two conceptually distinct approaches: general linear model (GLM) and functional connectivity analysis (independent component analysis, ICA). While the GLM analysis showed odor‐evoked activity in piriform cortex during olfactory stimulation as compared to pure sniffing, group ICA identified large‐scaled sensory processing networks recruited not only for odor processing but also for sniffing without odor stimulation. Task‐modulation of three networks was significantly correlated with scores of olfactory function (Hummel et al., 1997).
In both hyposmic and anosmic patients, we observed increased piriform cortex activation in response to olfactory stimulation as compared to pure sniffing. In general, piriform cortex activation during olfactory stimulation is well in line with a large number of previous studies showing the essential role of this region for olfactory processing (e.g., Lundström et al., 2011; Plailly et al., 2005; Seubert et al., 2013; Zelano et al., 2011). It is striking that the odor‐specific piriform cortex activation was not only present in hyposmic, but also in anosmic patients. Since the anosmic patients had no functional olfactory perception based on their Sniffin’ Sticks score, the observed piriform cortex activation in anosmics might be in line with evidence that odors can alter behavior and brain activity even if they are not consciously perceived (Arzi et al., 2014; Lorig, 2012; Smeets & Dijksterhuis, 2014; Sobel et al., 1999; Zucco et al., 2015). This finding suggests that the pathway from the olfactory epithelium to the piriform cortex might still be intact in these patients and that the olfactory dysfunction occurs after piriform processing. A recent study indicates that functional connections from piriform cortex to olfactory areas can be re‐established using olfactory training (Kollndorfer et al., 2014). Thus, one might speculate whether odor‐related activity in piriform cortex in anosmics is a prerequisite of susceptibility for such reorganization processes and whether the extent of this activity could play a role in disease prognosis and prediction of treatment success. It should be noted, that the observed piriform related activity in the odor > blank contrast could be driven by larger sniffs made in the odor condition. However, as patients received similar sniff instructions in both conditions, this seems unlikely.
In this study, we used two pleasant food‐related odors for stimulation. Food odors are biologically salient stimuli, and are processed differently in the brain compared to other types of odors (Boesveldt et al., 2010; Small, 2012). This might contribute to the perception of these odors remaining preserved, even in patients with severe olfactory dysfunction. This issue deserves to be further investigated in future studies comparing the processing of food‐related to non‐food‐related odors in olfactory dysfunction directly. Additionally, it is possible that some of the piriform cortex activation might be caused by trigeminal stimulation, although the odors were not selected to contain trigeminal properties, in contrast to, for example, Kollndorfer et al. (2015d). It is therefore recommended in future studies to thoroughly assess selected odors on trigeminal properties.
ICA revealed three networks which were recruited during the olfactory task and correlated with olfactory function scores: an olfactory, an occipital and a cerebellar network. These results support and extend a previous investigation on olfactory networks in participants with normal olfactory function (Karunanayaka et al., 2014). In this study, five functionally connected networks were involved in an olfactory task containing odor trials and no‐odor control trials. The two olfactory networks found in Karunanayaka et al. (2014) overlap to a large extent with the sensory processing network identified in our study, as they comprise traditional olfactory regions such as caudate, thalamus, putamen, and hippocampus. Interestingly, Karunanayaka et al. (2014) also identified a visual/occipital network modulated by their olfactory task, comprising parts of middle occipital gyrus. Notably, a network comprising the cerebellum was also identified but not described further due to the component selection criteria employed (Karunanayaka et al., 2014). Extending the findings of Karunanayaka et al. (2014), in the current study we were able to show that the task‐modulation of the networks during our olfactory paradigm was related to an external parameter, namely the scores participants achieved in an olfactory test. Thus, our results confirm the relevance of the sensory processing, occipital, and cerebellar networks by showing that the extent to which these networks are modulated by the olfactory task reflects olfactory function.
The sensory processing network identified in our study contained primarily regions previously associated with olfactory function (insula, thalamus, piriform cortex, cingulate cortex), but also further regions (superior temporal gyrus, superior frontal gyrus). Despite being not regarded as typical olfactory regions, an association of these regions with olfactory functions is in line with results of a recent a voxel‐based morphometry study investigating grey matter (GM) volume in anosmics (Peng et al., 2013). In this study, GM volume of superior temporal gyrus and superior frontal gyrus was decreased in anosmics compared to controls, possibly indicating an association of these regions with olfactory function.
The second network related to olfactory function in our study comprised occipital regions. Though visual input changed slightly when odor stimuli were released (the fixation cross changed color to signal the presentation of olfactory stimuli), this does not explain why recruitment of the network was related to scores achieved in the olfactory test. The occipital network comprised mainly the inferior and middle occipital gyrus, the middle temporal gyrus and the fusiform gyrus, areas not traditionally assumed to be main olfactory processing regions. However, a growing number of neuroimaging studies has reported activation of the visual cortex even during odor stimulation, particularly in olfactory identification and matching tasks (Kjelvik et al., 2012; Qureshy et al., 2000; Royet et al., 1999). Moreover, a decrease in grey matter volume of the fusiform gyrus, the middle temporal gyrus, and the middle occipital gyrus was demonstrated in previous voxel‐based morphometry studies in anosmics compared to healthy controls (Bitter et al., 2010; Peng et al., 2013), pointing to a possible role of these areas in olfactory processing. It has been suggested that during attempted identification of a smell, people might visualize the potential source of the odor (Jadauji et al., 2012). Thus, one might speculate whether patients scoring lower on the olfactory test in our study recruited the occipital network for visualization of the odor source less than patients achieving higher scores. This is in line with previous evidence that patients with olfactory deficits show a reduced olfactory imagery capacity (Flohr et al., 2014; Kollndorfer et al., 2015c). Furthermore, in a repetitive transcranial magnetic stimulation investigation, stimulation of the visual cortex led to improved odor discrimination performance as compared to sham stimulation, thus even pointing to a potential direct contribution of visual cortex to olfactory processing (Jadauji et al., 2012). The interconnection of the visual and olfactory system was also underlined by a recent study on olfactory‐visual conditioning (Hummel et al., 2017).
The recruitment of a cerebellar network also showed a correlation with olfactory function scores. Due to the requested button press to rate the stimuli during some (but not all) trials, a preparatory function of this network can be suspected. Moreover, previous neuroimaging studies demonstrating cerebellar activation in response to olfactory stimulation (e.g., Sobel et al., 1998b; Yousem et al., 1997; Zatorre et al., 2000) and with a reported impairment of olfactory function in patients with cerebellar lesions (Abele et al., 2003; Connelly et al., 2003). In particular, the cerebellum was suggested to be part of the “olfactomotor system” involved in the control of sniffing (Mainland & Sobel, 2006; Sobel et al., 1998a, 1998b). The particular importance of the cerebellar network for sniffing is further underlined by the correlation between network recruitment during pure sniffing trials and smelling function, with patients scoring higher in the olfactory test showing a higher task‐modulation of this network during sniffing. This result is well in line with an observed decreased functional connectivity of cerebellar regions in anosmic patients compared to normosmics during sniffing of odorless air (Kollndorfer et al., 2015a).
Interestingly, similarly to the findings observed for the cerebellar network, the recruitment of the sensory processing and occipital networks during pure sniffing trials was significantly correlated with olfactory function scores as well. Thus, our results support previous findings that sniffing alone can lead to activation of olfactory regions and extend them by showing that the extent of network recruitment is related to smelling function. Interestingly, in a previous olfactory study on persons with a normal olfactory function (Karunanayaka et al., 2014) olfactory network time courses were also task‐modulated in no‐odor control trials, as was the case in the present study. As discussed in Karunanayaka et al. (2014), this might reflect anticipation or expectation of odor stimulation by participants or reflect carry‐over effects from odor to nonodor trials.
In this study, we analyzed a relatively homogeneous sample of patients suffering from olfactory deficits, as we excluded those patients that might show neuronal changes unrelated to olfactory loss (e.g., hyposmia after head trauma or in the course of neurodegenerative diseases). Thus, although the correlative nature of our study impedes strong causal interpretations, we are confident that the recruitment of the functional networks can reflect the severity of olfactory deficits and not the effects of other potentially confounding factors. Still, an important question that could not be investigated in the present study is the relation between duration of olfactory disorder, brain activity patterns, and recruitment of functional networks, as duration of olfactory disorder was confounded with severity of the olfactory disorder within our population (Table 1). Duration of olfactory dysfunction therefore deserves to be investigated in further studies to gain more knowledge on the direction of the observed effects. Additionally, it was not possible to include patients with congenital anosmia in this study due to a low number of patients with this disorder in our population (n = 8). As Frasnelli et al. (2013) found that patients with congenital anosmia display fundamental changes in brain structure compared to healthy controls, it is recommended for studies further investigating the effects of duration of smell loss on functional networks to include patients with congenital disorders as well.
Our results indicate that even patients classified as anosmics based on olfactory testing scores can show activation in olfactory brain areas when stimulated with odors as compared to pure sniffing. Moreover, the recruitment of an olfactory, a cerebellar and an occipital network was related to olfactory function. Future studies might shed more light on the intriguing question whether such activation patterns might be predictive of disease progression or potential regain of olfactory function and of the success of treatment programs such as olfactory training.
ACKNOWLEDGMENTS
The authors want to thank all the MRI technicians from the Radiology department at Hospital Gelderse Vallei, Ede, The Netherlands, for their help with conducting the MRI scans. In particular, the authors want to thank Arjen Riemsma for his help in setting up the protocols. Furthermore, the Dr. Heinrich Jörg‐Stiftung, Karl‐Franzens‐Universität Graz, Austria, and the Styrian Provincial Government supported a research stay of JR in Wageningen. The authors declare no conflicts of interest.
Reichert JL, Postma EM, Smeets PAM, et al. Severity of olfactory deficits is reflected in functional brain networks—An fMRI study. Hum Brain Mapp. 2018;39:3166–3177. 10.1002/hbm.24067
REFERENCES
- Abele, M. , Riet, A. , Hummel, T. , Klockgether, T. , & Wullner, U. (2003). Olfactory dysfunction in cerebellar ataxia and multiple system atrophy. Journal of Neurology, 250(12), 1453–1455. [DOI] [PubMed] [Google Scholar]
- Arzi, A. , Rozenkrantz, L. , Holtzman, Y. , Secundo, L. , & Sobel, N. (2014). Sniffing patterns uncover implicit memory for undetected odors. Current Biology, 24(7), R263–R264. [DOI] [PubMed] [Google Scholar]
- Bitter, T. , Brüderle, J. , Gudziol, H. , Burmeister, H. P. , Gaser, C. , & Guntinas‐Lichius, O. (2010). Gray and white matter reduction in hyposmic subjects ‐ A voxel‐based morphometry study. Brain Research, 1347, 42–47. [DOI] [PubMed] [Google Scholar]
- Boesveldt, S. , Frasnelli, J. , Gordon, A. R. , & Lundström, J. N. (2010). The fish is bad: Negative food odors elicit faster and more accurate reactions than other odors. Biological Psychology, 84(2), 313–317. [DOI] [PubMed] [Google Scholar]
- Boesveldt, S. , Postma, E. , Boak, D. , Welge‐Luessen, A. , Schöpf, V. , Mainland, J. , … Duffy, V. (2017). Anosmia ‐ A clinical review. Chemical Senses, 42(7), 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brämerson, A. , Johansson, L. , Ek, L. , Nordin, S. , & Bende, M. (2004). Prevalence of olfactory dysfunction: The Skövde population‐based study. Laryngoscope, 114(4), 733–737. [DOI] [PubMed] [Google Scholar]
- Calhoun, V. D. , Adali, T. , Pearlson, G. D. , & Pekar, J. J. (2001). A method for making group inferences from functional MRI data using independent component analysis. Human Brain Mapping, 14(3), 140–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerf‐Ducastel, B. , & Murphy, C. (2003). FMRI brain activation in response to odors is reduced in primary olfactory areas of elderly subjects. Brain Research, 986(1–2), 39–53. [DOI] [PubMed] [Google Scholar]
- Connelly, T. , Farmer, J. M. , Lynch, D. R. , & Doty, R. L. (2003). Olfactory dysfunction in degenerative ataxias. Journal of Neurology, Neurosurgery, and Psychiatry, 74(10), 1435–1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croy, I. , Hoffmann, H. , Philpott, C. , Rombaux, P. , Welge‐Luessen, A. , Vodicka, J. , … Hummel, T. (2014a). Retronasal testing of olfactory function: An investigation and comparison in seven countries. European Archives of Oto‐Rhino‐Laryngology, 271, 1087–1095. [DOI] [PubMed] [Google Scholar]
- Croy, I. , Nordin, S. , & Hummel, T. (2014b). Olfactory disorders and quality of life ‐ An updated review. Chemical Senses, 39, 185–194. [DOI] [PubMed] [Google Scholar]
- Flohr, E. L. R. , Arshamian, A. , Wieser, M. J. , Hummel, C. , Larsson, M. , Mühlberger, A. , & Hummel, T. (2014). The fate of the inner nose: Odor imagery in patients with olfactory loss. Neuroscience, 268, 118–127. [DOI] [PubMed] [Google Scholar]
- Frasnelli, J. , Fark, T. , Lehmann, J. , Gerber, J. , & Hummel, T. (2013). Brain structure is changed in congenital anosmia. NeuroImage, 83, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Frasnelli, J. , Lundström, J. N. , Schöpf, V. , Negoias, S. , Hummel, T. , & Lepore, F. (2012). Dual processing streams in chemosensory perception. Frontiers in Human Neuroscience, 6, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, C. D. , & Sigman, M. (2007). Brain states: Top‐down influences in sensory processing. Neuron, 54(5), 677–696. [DOI] [PubMed] [Google Scholar]
- Griffioen‐Roose, S. , Smeets, P. A. M. , van den Heuvel, E. , Boesveldt, S. , Finlayson, G. , & de Graaf, C. (2014). Human protein status modulates brain reward responses to food cues 1 – 3. American Journal of Clinical Nutrition, 100(1), 113–122. [DOI] [PubMed] [Google Scholar]
- Himberg, J. , Hyvärinen, A. , & Esposito, F. (2003). Validating the independent components of neuroimaging time‐series via clustering and visualization. [DOI] [PubMed]
- Hummel, T. , Kobal, G. , Gudziol, H. , & Mackay‐Sim, A. (2007). Normative data for the “Sniffin’ Sticks” including tests of odor identification, odor discrimination, and olfactory thresholds: An upgrade based on a group of more than 3,000 subjects. European Archives of Oto‐Rhino‐Laryngology, 264(3), 237–243. [DOI] [PubMed] [Google Scholar]
- Hummel, T. , Sekinger, B. , Wolf, S. R. , Pauli, E. , & Kobal, G. (1997). Sniffin” sticks’. Olfactory performance assessed by the combined testing of odor identification, odor discrimination and olfactory threshold. Chemical Senses, 22(1), 39–52. [DOI] [PubMed] [Google Scholar]
- Hummel, T. , Fark, T. , Baum, D. , Warr, J. , Hummel, C. B. , & Schriever, V. A. (2017). The rewarding effect of pictures with positive emotional connotation upon perception and processing of pleasant odors—An FMRI study. Frontiers in Neuroanatomy, 11, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadauji, J. B. , Djordjevic, J. , Lundstrom, J. N. , & Pack, C. C. (2012). Modulation of olfactory perception by visual cortex stimulation. Journal of Neuroscience, 32(9), 3095–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karunanayaka, P. , Eslinger, P. J. , Wang, J.‐L. , Weitekamp, C. W. , Molitoris, S. , Gates, K. M. , … Yang, Q. X. (2014). Networks involved in olfaction and their dynamics using independent component analysis and unified structural equation modeling. Human Brain Mapping, 35(5), 2055–2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjelvik, G. , Evensmoen, H. R. , Brezova, V. , & Håberg, A. K. (2012). The human brain representation of odor identification. Journal of Neurophysiology, 108(2), 645–657. [DOI] [PubMed] [Google Scholar]
- Kobal, G. , Hummel, T. , Sekinger, B. , Barz, S. , Roscher, S. , & Wolf, S. (1996). Sniffin’ sticks”: Screening of olfactory performance. Rhinology, 34(4), 222–226. [PubMed] [Google Scholar]
- Kollndorfer, K. , Fischmeister, F. P. S. , Kowalczyk, K. , Hoche, E. , Mueller, C. A. , Trattnig, S. , & Schöpf, V. (2015a). Olfactory training induces changes in regional functional connectivity in patients with long‐term smell loss. NeuroImage: Clinical, 9, 401–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollndorfer, K. , Jakab, A. , Mueller, C. A. , Trattnig, S. , & Schöpf, V. (2015b). Effects of chronic peripheral olfactory loss on functional brain networks. Neuroscience, 310, 589–599. [DOI] [PubMed] [Google Scholar]
- Kollndorfer, K. , Kowalczyk, K. , Frasnelli, J. , Hoche, E. , Unger, E. , Mueller, C. A. , … Schöpf, V. (2015c). Same same but different. Different trigeminal chemoreceptors share the same central pathway. PLoS One, 10, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollndorfer, K. , Kowalczyk, K. , Hoche, E. , Mueller, C. A. , Pollak, M. , Trattnig, S. , & Schöpf, V. (2014). Recovery of olfactory function induces neuroplasticity effects in patients with smell loss. Neural Plasticity, 2014, 1. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollndorfer, K. , Kowalczyk, K. , Nell, S. , Krajnik, J. , Mueller, C. A. , & Schöpf, V. (2015d). The inability to self‐evaluate smell performance. How the vividness of mental images outweighs awareness of olfactory performance. Frontiers in Psychology, 6, 627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorig, T. S. (2012). Beyond self‐report: Brain imaging at the threshold of odor perception. Chemosensory Perception, 5(1), 46–54. [Google Scholar]
- Lötsch, J. , Ultsch, A. , Eckhardt, M. , Huart, C. , Rombaux, P. , & Hummel, T. (2016). Brain lesion‐pattern analysis in patients with olfactory dysfunctions following head trauma. NeuroImage: Clinical , 11, 99–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundström, J. N. , Boesveldt, S. , & Albrecht, J. (2011). Central processing of the chemical senses: An overview. ACS Chemical Neuroscience, 2(1), 5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainland, J. , & Sobel, N. (2006). The sniff is part of the olfactory percept. Chemical Senses, 31(2), 181–196. [DOI] [PubMed] [Google Scholar]
- Mazaika, P. K. , Hoeft, F. , Glover, G. H. , & Reiss, A. L. (2009). Methods and software for fMRI analysis of clinical subjects. NeuroImage, 47, S58. [Google Scholar]
- Mueller, C. , Kallert, S. , Renner, B. , & Stiassny, K. (2003). Quantitative assessment of gustatory function in a clinical context using impregnated” taste strips. Rhinology, 41, 2–6. [PubMed] [Google Scholar]
- Patel, Z. M. (2017). The evidence for olfactory training in treating patients with olfactory loss. Current Opinion in Otolaryngology &Amp; Head and Neck Surgery, 25(1), 43–46. [DOI] [PubMed] [Google Scholar]
- Peng, P. , Gu, H. , Xiao, W. , Si, L. F. , Wang, J. F. , Wang, S. K. , … Wei, Y. X. (2013). A voxel‐based morphometry study of anosmic patients. British Journal of Radiology, 86(1032), 20130207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plailly, J. , Bensafi, M. , Pachot‐Clouard, M. , Delon‐Martin, C. , Kareken, D. A. , Rouby, C. , … Royet, J. P. (2005). Involvement of right piriform cortex in olfactory familiarity judgments. NeuroImage, 24(4), 1032–1041. [DOI] [PubMed] [Google Scholar]
- Qureshy, A. , Kawashima, R. , Babar Imran, M. , Sugiura, M. , Goto, R. , Okada, K. , … Babar Im‐Ran, M. (2000). Functional mapping of human brain in olfactory processing: A PET study. Journal of Neurophysiology, 84(3), 1656–1666. [DOI] [PubMed] [Google Scholar]
- Rawal, S. , Hoffman, H. J. , Bainbridge, K. E. , Huedo‐Medina, T. B. , & Duffy, V. B. (2016). Prevalence and risk factors of self‐reported smell and taste alterations: Results from the 2011–2012 US national health and nutrition examination survey (NHANES). Chemical Senses, 41(1), 69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichert, J. L. , Ninaus, M. , Schuehly, W. , Hirschmann, C. , Bagga, D. , & Schöpf, V. (2017). Functional brain networks during picture encoding and recognition in different odor contexts. Behavioural Brain Research, 333, 98–108. [DOI] [PubMed] [Google Scholar]
- Reichert, J. L. , & Schöpf, V. (2017). Olfactory loss and regain: Lessons for neuroplasticity. Neuroscience, 107385841770391. [DOI] [PubMed] [Google Scholar]
- Royet, J. P. , Koenig, O. , Gregoire, M. C. , Cinotti, L. , Lavenne, F. , Le Bars, D. , … Froment, J. C. (1999). Functional anatomy of perceptual and semantic processing for odors. Journal of Cognitive Neuroscience, 11(1), 94–17. [DOI] [PubMed] [Google Scholar]
- Seubert, J. , Freiherr, J. , Frasnelli, J. , Hummel, T. , & Lundström, J. N. (2013). Orbitofrontal cortex and olfactory bulb volume predict distinct aspects of olfactory performance in healthy subjects. Cerebral Cortex (New York, N.Y. : 1991), 23(10), 2448–2456. [DOI] [PubMed] [Google Scholar]
- Small, D. M. (2012). Flavor is in the brain. Physiology &Amp; Behavior, 107(4), 540–552. [DOI] [PubMed] [Google Scholar]
- Smeets, M. , & Dijksterhuis, G. (2014). Smelly primes – when olfactory primes do or do not work. Frontiers in Psychology, 5, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel, N. , Prabhakaran, V. , Desmond, J. E. , Glover, G. H. , Goode, R. L. , Sullivan, E. V. , & Gabrieli, J. D. (1998a). Sniffing and smelling: Separate subsystems in the human olfactory cortex. Nature, 392, 282–286. [DOI] [PubMed] [Google Scholar]
- Sobel, N. , Prabhakaran, V. , Hartley, C. A. , Desmond, J. E. , Zhao, Z. , Glover, G. H. , … Sullivan, E. V. (1998b). Odorant‐induced and sniff‐induced activation in the cerebellum of the human. Journal of Neuroscience, 18, 8990–9001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel, N. , Prabhakaran, V. , Hartley, C. A. , Desmond, J. E. , Glover, G. H. , Sullivan, E. V. , & Gabrieli, J. D. E. (1999). Blind smell: Brain activation induced by an undetected air‐borne chemical. Brain, 122(2), 209–217. [DOI] [PubMed] [Google Scholar]
- Sorokowska, A. , Albrecht, E. , Haehner, A. , & Hummel, T. (2015). Extended version of the “Sniffin ’ Sticks” identification test : Test – retest reliability and validity. Journal of Neuroscience Methods, 243, 111–114. [DOI] [PubMed] [Google Scholar]
- Temmel, A. F. P. , Quint, C. , Schickinger‐Fischer, B. , Klimek, L. , Stoller, E. , & Hummel, T. (2002). Characteristics of olfactory disorders in relation to major causes of olfactory loss. Archives of Otolaryngology–Head & Neck Surgery, 128(6), 635–641. [DOI] [PubMed] [Google Scholar]
- Tzourio‐Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, … Joliot M. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single‐subject brain. Neuroimage. 2002;15(1), 273–289. [DOI] [PubMed] [Google Scholar]
- Vedaei, F. , Fakhri, M. , Harirchian, M. H. , Firouznia, K. , Lotfi, Y. , & Ali Oghabian, M. (2013). Methodological considerations in conducting an olfactory fMRI study. Behavioural Neurology, 27(3), 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. , Eslinger, P. J. , Doty, R. L. , Erin, K. , Grunfeld, R. , Sun, X. , … Yang, Q. X. (2010). Olfactory deficit detected by fMRI in early Alzheimer's Disease. Brain Research, 1357, 184–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welge‐Lüssen, A. (2009). Ageing, neurodegeneration, and olfactory and gustatory loss. B‐ENT, 5(Suppl 13), 129–132. [PubMed] [Google Scholar]
- Welge‐Lüssen, A. , Wattendorf, E. , Schwerdtfeger, U. , Fuhr, P. , Bilecen, D. , Hummel, T. , & Westermann, B. (2009). Olfactory‐induced brain activity in Parkinson's disease relates to the expression of event‐related potentials: A functional magnetic resonance imaging study. Neuroscience, 162(2), 537–543. [DOI] [PubMed] [Google Scholar]
- Yousem, D. M. , Williams, S. C. , Howard, R. O. , Andrew, C. , Simmons, A. , Allin, M. , … Doty, R. L. (1997). Functional MR imaging during odor stimulation: Preliminary data. Radiology, 204(3), 833–838. [DOI] [PubMed] [Google Scholar]
- Yousem, D. M. , Geckle, R. J. , Bilker, W. B. , Kroger, H. , & Doty, R. L. (1999). Posttraumatic smell loss: Relationship of psychophysical tests and volumes of the olfactory bulbs and tracts and the temporal lobes. Academic Radiology, 6(5), 264–272. [DOI] [PubMed] [Google Scholar]
- Zatorre, R. J. , Jones‐Gotman, M. , & Rouby, C. (2000). Neural mechanisms involved in odor pleasantness and intensity judgments. Neuroreport, 11(12), 2711–2716. [DOI] [PubMed] [Google Scholar]
- Zelano, C. , Mohanty, A. , & Gottfried, J. A. (2011). Olfactory predictive codes and stimulus templates in piriform cortex. Neuron, 72(1), 178–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoon, H. F. A. , Graaf, C. , & De, Boesveldt, S. (2016). Food odours direct specific appetite. Foods, 5(1), 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucco, G. M. , Priftis, K. , & Stevenson, R. J. (2015). From blindsight to blindsmell: A mini review. Translational Neuroscience, 6(1), 8–12. [DOI] [PMC free article] [PubMed] [Google Scholar]


