Abstract
Tensor‐based morphometry (TBM) performed using T1‐weighted images (T1WIs) is a well‐established method for analyzing local morphological changes occurring in the brain due to normal aging and disease. However, in white matter regions that appear homogeneous on T1WIs, T1W‐TBM may be inadequate for detecting changes that affect specific pathways. In these regions, diffusion tensor MRI (DTI) can identify white matter pathways on the basis of their different anisotropy and orientation. In this study, we propose performing TBM using deformation fields constructed using all scalar and directional information provided by the diffusion tensor (DTBM) with the goal of increasing sensitivity in detecting morphological abnormalities of specific white matter pathways. Previously, mostly fractional anisotropy (FA) has been used to drive registration in diffusion MRI‐based TBM (FA‐TBM). However, FA does not have the directional information that the tensors contain, therefore, the registration based on tensors provides better alignment of brain structures and better localization of volume change. We compare our DTBM method to both T1W‐TBM and FA‐TBM in investigating differences in brain morphology between patients with complicated hereditary spastic paraplegia of type 11 (SPG11) and a group of healthy controls. Effect size maps of T1W‐TBM of SPG11 patients showed diffuse atrophy of white matter. However, DTBM indicated that atrophy was more localized, predominantly affecting several long‐range pathways. The results of our study suggest that DTBM could be a powerful tool for detecting morphological changes of specific white matter pathways in normal brain development and aging, as well as in degenerative disorders.
Keywords: DTBM, deformation‐based morphometry, diffusion, diffusion tensor imaging, diffusion tensor MRI, hereditary spastic paraplegia, spastic paraplegia of type 11, tensor‐based morphometry
1. INTRODUCTION
Across the lifespan in health and in disease, brain morphology undergoes remarkable changes. To assess interindividual and group differences in brain anatomy from MRI scans, often the image data of each subject are registered into a common space or template. The deformation fields that map individual images from their native space to the common space can then be used as a measure of difference in brain morphology. This approach is generally known as deformation‐based morphometry. When the spatial derivatives of the deformation fields (the Jacobian) are used, the approach is called tensor‐based morphometry (TBM; Ashburner et al., 1998; Ashburner & Friston, 2004; Davatzikos et al., 1996). An important feature of the deformation field is the determinant of the Jacobian (J). J encodes the volume of the deformed unit‐cube after registration, therefore it is a measure of local expansion or contraction of volume. In imaging, a voxel can be considered as a unit‐cube. A J value of 1 indicates no change in volume, J < 1 indicates local contraction (i.e., tissue loss), and J > 1 indicates local expansion (i.e., growth), therefore, J can be used as a method to detect local volume change in development or disease. Indeed, TBM has been used to study normal brain development and many brain disorders including Alzheimer's disease, HIV/AIDS, Huntington's disease, and schizophrenia (Chiang et al., 2007; Hua et al., 2008; Kipps et al., 2005; Thompson et al., 2000). TBM has been predominantly performed using structural MRI images, in particular T1‐weighted images (T1WIs) to drive the registration algorithm, as they provide good delineation between white matter, gray matter, and cerebrospinal fluid (CSF). However, it is important to consider that the signal intensity within white matter appears relatively homogenous in T1WIs making it difficult to detect morphological changes of specific white matter pathways contained in large white matter regions.
Diffusion tensor imaging (DTI; Basser, Mattiello, & LeBihan, 1994) is an MRI modality that is particularly suitable for identifying individual white matter pathways (Pierpaoli, Jezzard, Basser, Barnett, & Di Chiro, 1996). DTI provides information about the principal direction of diffusion which is locally collinear with the orientation of major fiber bundles, as well as scalar metrics, such as fractional anisotropy (FA) and the mean diffusivity (MD; i.e., Trace of the diffusion tensor/3; Basser & Pierpaoli, 1996). TBM has been applied to diffusion MRI data in limited cases. However, mainly the scalar maps derived from DTI, such as FA or low b‐value DWIs were used to drive the registration (Oishi et al., 2011; Pagani, Horsfield, Rocca, & Filippi, 2007; Verma et al., 2005) with the exception of work by Studholme (2008), where a weighted combination of diffusion tensor elements and T1WIs were used. Using the directional information provided by the diffusion tensor enables a better identification of individual white matter pathways compared to using scalar diffusion metrics, such as FA or T1WIs and T2‐weighted images (T2WIs; Pajevic & Pierpaoli, 1999). For example, the cingulum bundle (CB) can easily be differentiated from the corpus callosum (CC) or the corticospinal tract (CST) from the transverse pontine fibers in the directionally encoded color (DEC) maps (Pajevic & Pierpaoli, 1999), while the demarcation between these pathways is not easy to assess with FA, T1WIs, or T2WIs.
Alignment of diffusion tensor images is a more involved process compared to the alignment of scalar images as the tensors must be reoriented to be consistent with the reorientation of anatomical structures. There has been extensive work on performing proper alignment of diffusion tensor images, for example, using the deviatoric of the diffusion tensor and optimizing tensor orientation explicitly (Zhang et al., 2007), and using the full diffusion tensor (Irfanoglu et al., 2016). However, previous work has concentrated on the improvement of registration and subsequent voxelwise analysis of diffusion derived metrics and not on the TBM of deformation fields themselves. In this study, we propose to use diffusion tensor data to drive the registration, taking advantage of both DTI scalar and vectorial information and perform TBM on these deformation fields. We hypothesize that TBM based on deformation fields obtained by registration of diffusion tensors (DTBM) will be more sensitive than TBM based on scalar images, such as T1WIs (T1W‐TBM) and FA (FA‐TBM) in detecting morphometric changes in specific white matter pathways. We used all the three TBM methods, DTBM, T1W‐TBM, and FA‐TBM, to assess their ability to characterize morphometric abnormalities with respect to controls in a group of patients with spastic paraplegia of type 11 (SPG11), a rare disorder that is well defined genetically, and has certain disease characteristics involving atrophy of white matter, making it particularly suitable for testing our hypothesis.
Spastic paraplegia of type 11 is a subtype of hereditary spastic paraplegia (HSP), a group of inherited disorders characterized by progressive spasticity and lower limb weakness produced by abnormalities in the protein spatacsin caused by a mutation in the SPG11 gene. Although the function of spatacsin is not completely clear, current evidence suggests it may be involved in anterograde vesicle trafficking and axonal transport (Pérez‐Brangulí et al., 2014), both of which are expected to play a crucial role in axonal maintenance, in particular for long‐range white matter pathways. SPG11 is therefore an appropriate candidate for assessing the sensitivity of TBM methods as multiple white matter pathways including commissural, association, and projection pathways are affected (Cao et al., 2013; Chen et al., 2008; França et al., 2012; Garaci et al., 2014). A common MRI finding is a thin CC, which is visually detectable (Cao et al., 2013; Fink, 2013; Stevanin et al., 2007). In addition, a common documented pathological feature of HSP is the axonal degeneration of the CST (motor pathway) along the medulla and spinal cord (Deluca, Ebers, & Esiri, 2004; McDermott, White, Bushby, & Shaw, 2000; Salinas, Proukakis, Crosby, & Warner, 2008; Wharton et al., 2003; White et al., 2000). We hypothesized that DTBM will have an advantage in detecting CST atrophy, especially at the level of the brainstem where DTI provides additional information (these regions appear homogenous in the T1WIs). Another region of interest is the centrum semiovale where multiple long‐range pathways are located, DTBM might be able to detect whether these white matter tracts are differentially affected as this region also appears homogenous in the T1WIs. Moreover, if the role recently proposed for spatacsin by Pérez‐Brangulí et al. (2014) in SPG11 involves more selective loss of long‐range fibers, DTBM should be able to detect a pattern of white matter atrophy that affects preferentially selected white matter pathways.
2. MATERIALS AND METHODS
2.1. Subjects
Twenty‐four healthy volunteers (mean age of 35 and standard deviation of six years; 15 female and 9 male) with no history of neurological disorders and normal MRI, and four subjects diagnosed with SPG11 (mean age of 32 and standard deviation of three years; four female) were included in this study. The study was approved by the Ethics Committee and written informed consent was obtained from all subjects.
2.2. Image acquisition
All participants were scanned on a Philips 3T system with a 32‐channel head coil. The DTI data were acquired with a single‐shot spin‐echo echo‐planar imaging (EPI) sequence (repetition time [TR]: 4,700 ms, echo time [TE]: 80 ms, 80 slices, acceleration factor: 2 with an isotropic voxel size of 2.2 × 2.2 × 2.2 mm3). A multishell DTI protocol composed of eight nondiffusion‐weighted images along with 15 volumes with b‐value of 300 s/mm2 and 53 volumes with b‐value of 1,100 s/mm2. Additionally, MPRAGE 3D T1WIs (sequence parameters: TR: 8.2 ms, TE: 3.8 ms, voxel size: approximately 1 × 1 × 1 mm3) and T2WIs (sequence parameters: TR: 3,000 ms, TE: 100 ms, voxel size: approximately 1.5 × 1.5 × 1.7 mm3) were obtained.
2.3. Diffusion‐weighted image processing
Diffusion‐weighted volumes were processed with the diffprep module of the TORTOISE software package1 (Irfanoglu, Nayak et al., 2017; Pierpaoli et al., 2010). The processing steps included: reduction of the effects of motion and eddy current distortions with appropriate rotations to the b‐matrix (Rohde, Barnett, Basser, Marenco, & Pierpaoli, 2004), correction of EPI distortions by nonlinear B‐spline registration to the individual's undistorted structural T2WI (Wu et al., 2008), and reorientation of all volumes to a common standard orientation with the midsagittal plane of the image separating the two hemispheres and the intersection of the anterior and posterior commissures with the sagittal plane lying on the same axial slice (midsagittal and AC–PC alignment). All corrected DWIs were systematically inspected for the presence of uncorrected motion. A DWI volume was removed if it showed uncorrected motion. Subsequently, robust estimation by outlier rejection tensor estimation (RESTORE; Chang, Jones, & Pierpaoli, 2005; Chang, Walker, & Pierpaoli, 2012) of the diffcalc module of TORTOISE package (Pierpaoli et al., 2010) was used to estimate the diffusion tensor.
2.4. Spatial normalization and construction of Jacobian maps
In general, an average template is a 3D image that should represent the average of features (e.g., size, shape, and composition) of the imaged anatomical organ in the population of interest. Spatial normalization is necessary for both constructing the average template and assessing morphological differences between individual subjects and the template. We used two software programs for spatial normalization. For T1W images and FA maps, we used the well‐established ANTS software (Avants et al., 2010, 2011). For DTI spatial normalization we used the recently developed DR‐TAMAS software (Irfanoglu et al., 2016).
DR‐TAMAS is a diffeomorphic diffusion tensor‐based registration method that uses all scalar and vectorial tensor features (anisotropy, Trace, eigenvectors) in its cost function, achieving good overall quality of registration in white matter, gray matter, and CSF regions (Irfanoglu et al., 2016). For the purpose of template building, at each iteration, we added the constraint that the sum of the displacement fields from the template to each individual image is zero at each voxel location to ensure the computed template represents the average morphology of the population (Irfanoglu, Sadeghi et al., 2017). We built modality‐specific control templates using ANTS and DR‐TAMAS software with the symmetric normalization (SyN) diffeomorphic transformation model option proposed by Avants, Epstein, Grossman, and Gee (2008). SyN transformation model has been shown to be powerful and fast (Klein et al., 2009; Murphy et al., 2011).
For each modality (T1W, FA, and DTI), individual subjects were registered to the control template, and the corresponding transformations (combined affine and nonlinear transformations) were used to compute the modality‐specific log of determinant of Jacobian (LogJ) maps for each subject. To test the hypothesis that long‐range fibers were more selectively affected, deterministic tractography (Mori & van Zijl, 2002; Wang, Benner, Sorensen, & Wedeen, 2007) performed on the DTI average template of the control subjects was used to assess which pathways pass through regions that showed localized atrophy (negative values in the LogJ maps) in SPG11 patients. The angular threshold for terminating streamline propagation was set to 20° as this has been shown to yield relatively higher specificity in the anatomical accuracy of pathways visualized using diffusion tractography (Thomas et al., 2014). The extracted tracts were identified and labeled according to the terminology used by Catani and de Schotten (2012).
2.5. Statistical analysis
Permutation testing was performed to determine whether the LogJ maps of SPG11 were significantly different from the control. FSL randomize software was used to determine the p values (Winkler, Ridgway, Webster, Smith, & Nichols, 2014) and corrected for multiple comparisons using family‐wise error rate of 5% (p < .05). In addition, effect size maps were computed using the following formula: ([mean of patients] – [mean of controls])/pooled standard deviation. As the two groups are dissimilar in size, we used pooling of weighted standard deviation such that each group's standard deviation is weighted by its sample size (Hedges, 1981).
3. RESULTS
Figure 1 shows voxelwise maps of the effect size for LogJ maps obtained from T1W‐TBM as it is typically performed, as well as FA‐TBM which also uses scalar images for registration, and our proposed DTBM method which uses tensor data for registration. The axial slices show effect size for LogJ maps at different rostrocaudal brain levels and, being scaled with the same window, allow comparison of these three different approaches. Negative values (darker regions) in the effect size maps indicate areas that are smaller in the patients with respect to the controls, and positive values (brighter regions) indicate regions that are larger in the patients compared to the controls. Note that the scale is from negative seven to seven, where an effect size of 0.8 is considered a large effect size (Cohen, 1988). As one of the main characteristics of HSP is progressive weakness and spasticity of the lower limbs, it is important to examine the results specifically with regard to the changes in the apparent size of the CST in the patient group. Figure 1a–c shows the CST at the level of the medulla where a sensitive method should be able to detect atrophy of the CST, which is well documented histologically in SPG11 (Deluca et al., 2004; McDermott et al., 2000; Salinas et al., 2008; Wharton et al., 2003; White et al., 2000). Atrophy is clearly evident for data processed with DTBM, while the T1W‐TBM maps are unremarkable at this level. This is probably caused by the lack of contrast in this region in the T1W images. FA‐TBM maps show some CST atrophy, but less clearly than DTBM.
Figure 1.
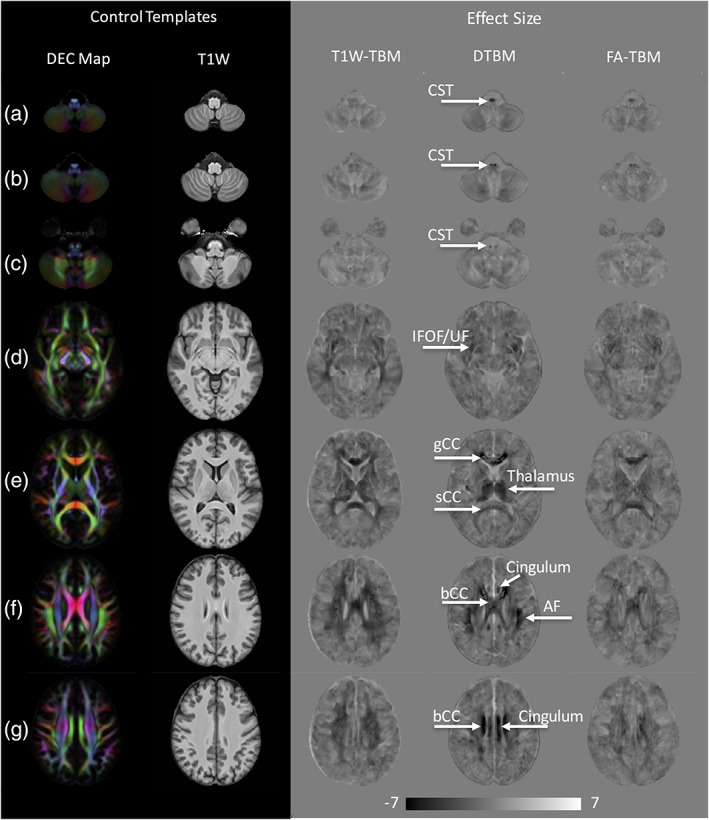
Control templates and effect size of LogJ maps for T1W‐TBM, DTBM, and FA‐TBM at seven different axial slices (a–g) are shown. Dark voxels in the effect size images represent regions where structures in the patient group are smaller than the control group (regions of atrophy or hypoplasia), whereas the bright regions, such as CSF spaces and ventricles, are areas that are larger in the patient group compared to the control group. Fiber tracts that show large effect size in DTBM are labeled. AF = arcuate fasciculus, bCC = body of CC, CST = corticospinal tract; gCC = fibers from the genu of the CC; IFOF = inferior fronto‐occipital fasciculus; sCC = fibers from the splenium of the CC; UF = uncinate fasciculus. The brain is presented in radiological convention with the right hemisphere on the left of the image
Figure 1d shows atrophy of additional white matter pathways projecting along the extreme capsule. This region has been described as an area, where several long‐range white matter pathways converge (Maier‐Hein et al., 2017) making it difficult to distinguish whether the atrophy is specific to the inferior fronto‐occipital fasciculus (IFOF) or the uncinate fasciculus (UF). Nonetheless, all three approaches identify atrophy in this region, with T1W‐TBM also indicating atrophy in the posterior region of the brain.
Next, we looked at the effect size maps at the level of CC (Figure 1e). A common finding in conventional MRI data of SPG11 patients is thin CC. Effect size maps of all three methods (DTBM, FA‐TBM, and T1W‐TBM) also indicate a thin CC for the SPG11 patients, however, the magnitude and extent of the effect is larger in the DTBM compared to the T1W‐TBM and FA‐TBM. Also, of note is the indication of atrophy of the thalamus and enlargement of ventricles at this level (Figure 1e), which is evident in DTBM and T1W‐TBM, but not in FA‐TBM, which fail to differentiate the anterior horn of lateral ventricles and the caudate nuclei. This is not surprising given the low FA of both CSF and the gray matter, there is no contrast between these two structures to drive the registration in FA‐TBM. Both T1W‐TBM and DTBM are much more effective in showing the specific enlargement of the lateral ventricles.
Figure 1f,g shows axial slices at the level of the centrum semiovale, which contains association, commissural, and projection fibers. The centrum semiovale appears as a homogenous white matter region in the T1W images, however, multiple fiber bundles are evident on the DEC maps. The effect size maps of Figure 1f,g show that white matter architecture along the centrum semiovale is greatly altered in the SPG11 patient group. Effect size maps of T1W‐TBM show diffuse atrophy in this region, whereas, DTBM maps help identify localized white matter pathways that are more selectively affected. Cingulum, CC, and arcuate fasciculus (AF) are among the white matter pathways that are severely affected, as indicated in the DTBM maps and to a much lesser degree in the FA‐TBM.
Figure 2 shows the results of the statistical voxelwise analysis of LogJ maps. Regions in red correspond to statistically significant white matter atrophy in SPG11 patients. All three TBM approaches show some areas of significant volumetric reduction in SPG11 patients compared to the control group, but the spatial localization and the extent of the regions are different for the different methods. DTBM atrophy is localized and predominantly affects several long‐range white matter pathways. These specific white matter pathways (Figure 3) can be traced by following dark regions (indicative of volume loss) in the effect size maps (Figure 1). These white matter tracts include commissural pathways, association pathways (most prominently the AF, but also the IFOF, UF, and CB), and projection pathways (CST). In addition to white matter regions, significant atrophy was also detected in gray matter regions, such as the thalamus (Figure 2e). The spatial extent of the atrophy detected by T1W‐TBM is relatively large but less localized to specific white matter bundles. FA‐TBM shows the least spatial extent of atrophy. Interestingly, only DTBM detects significant atrophy of the CST.
Figure 2.
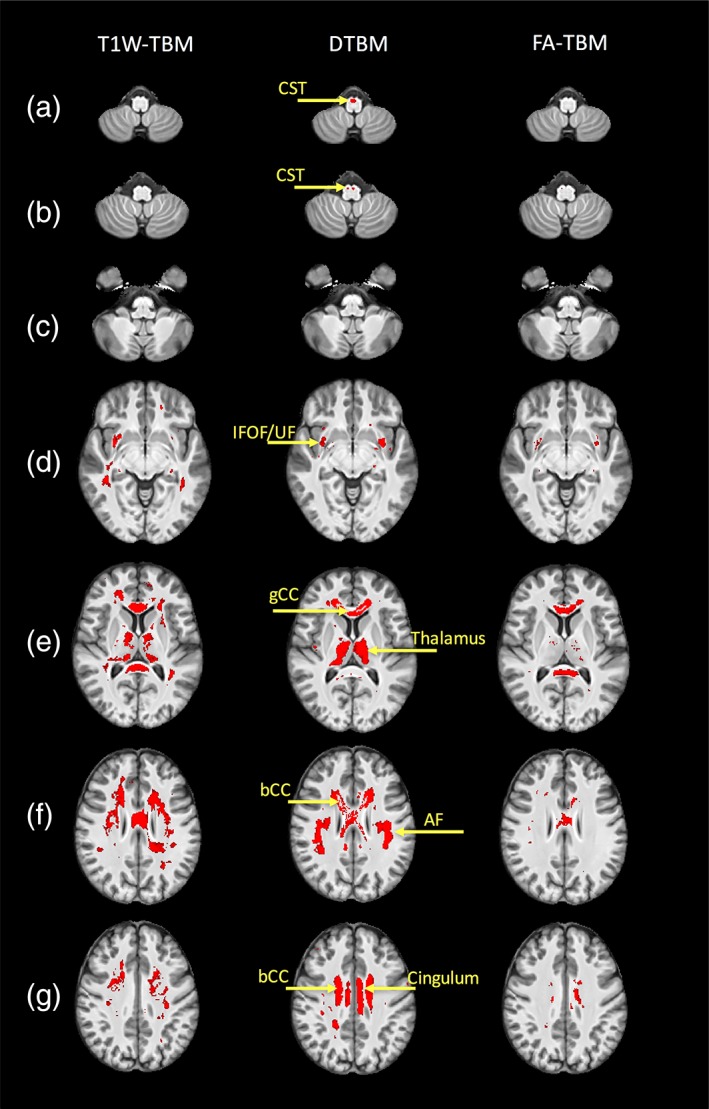
Regions of statistically significant atrophy (red regions) in the SPG11 patients compared to the controls using the three different approaches: T1W–TBM (left column), DTBM (middle column), and FA–TBM (right column). Significance map is overlaid on the control T1W template. Fiber tracts that show statistically significant volumetric reduction are: AF = arcuate fasciculus; bCC = body of CC; CST = corticospinal tract; gCC = fibers from the genu of the CC; IFOF = inferior fronto‐occipital fasciculus; UF = uncinate fasciculus. The brain is presented in radiological convention with the right hemisphere on the left of the image
Figure 3.
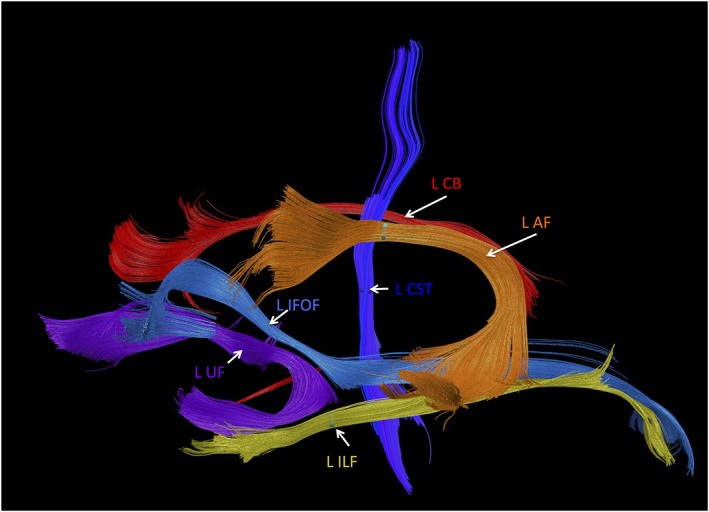
Fiber tracts that pass through regions of atrophy detected by DTBM in the left hemisphere. AF = arcuate fasciculus; CB = cingulum bundle; CST = corticospinal tract; IFOF = inferior fronto‐occipital fasciculus; ILF = inferior longitudinal fasciculus; UF = uncinate fasciculus. Remarkably, these are all long‐range fiber tracts
In addition to group analysis, we looked at the brain morphology at the single subject level by generating z‐score maps for each of the SPG11 subjects (see [Link], [Link], [Link]). DTBM showed a pattern of atrophy that is symmetric between left and right hemispheres and consistent across subjects (Supporting Information Figure S1). For example, Supporting Information Figure S1f,g shows different fiber tracts in centrum semiovale, which are consistently detected for each subject, and the magnitude of the z‐scores is larger compared to T1W‐TBM and FA‐TBM. Atrophy of CST was detected with a z‐score of >4 in three of the four SPG subjects (Supporting Information Figure S1a,b), but was not detected by T1W‐TBM and only detected in two subject with a lower z‐score by FA‐TBM. Similar to the group comparison, T1W‐TBM showed diffuse white matter atrophy at the individual level, however, the regions of atrophy were not as consistent among SPG11 subjects as the regions identified by DTBM (Supporting Information Figure S2). FA‐TBM showed less atrophy overall compared to the other two methods (Supporting Information Figure S3).
4. DISCUSSION
In this study, we proposed a novel way of performing TBM that is driven by scalar and vectorial information of the diffusion tensor. DTBM offers multiple advantages over the scalar‐based TBM methods of T1W‐TBM and FA‐TBM, including the ability to localize patterns of volume change to specific fiber pathways within white matter regions that appear homogenous in T1W images. Additionally, DTBM was found to detect volume changes with higher sensitivity, as indicated by the greater magnitude of its effect size. Finally, DTBM was able to reveal volume abnormalities that were invisible using other TBM methods in tracts with known involvement in HSP, such as the CST (Deluca et al., 2004; McDermott et al., 2000; Salinas et al., 2008; Wharton et al., 2003; White et al., 2000).
We chose to evaluate brains of SPG11 patients to evaluate the utility of our proposed method, because SPG11 shows evident white matter atrophy in the CC, which could imply atrophy of other long‐range white matter pathways (França et al., 2012), in addition to diffusion anisotropy abnormalities detected using DTI (França et al., 2012; Garaci et al., 2014). While both morphometric and microstructural alterations are implicated in this disorder, it is important to emphasize that they can have different etiology and consequences, and therefore the primary goal of this study was to evaluate morphometric analysis tools to complement the analysis of DTI scalar quantities, such as FA, that have already been described in the literature.
Given the disorder's rarity, most previous studies including our study are limited to a small number of patients, but anatomical abnormalities that accompany the disorder are quite consistent across subjects and large in magnitude. For example, in several long‐range white matter pathways we found an effect size for DTBM that was nearly an order of magnitude higher than what is considered a large effect size. The prominence of abnormalities, along with a suitable number of controls, allowed the generation of z‐score maps for assessing brain morphology at a single subject level (see [Link], [Link], [Link]). Taken together, the findings of this study support the use of DTBM to provide morphometric metrics that are not available using other approaches.
4.1. Potential of DTBM in characterizing CNS disorders and its relationship to DTI
In this study, we have shown that DTBM provides useful information about white matter changes in SPG11 patients. On a more general level, one could ask what role DTBM could play in characterizing other CNS disorders and how its use could be integrated into a more classical voxel‐based analyses of diffusion metrics, such as FA and MD. While voxelwise analysis of FA or MD offers insights about changes in microstructure and architecture (e.g., intravoxel orientational arrangement of fibers) within a structure of interest, DTBM analysis offers insights about changes in the size of a given structure. In some disorders, both the microstructure and the size of the fiber bundle might be affected, which is likely the case for SPG11. Previous studies have indeed found diffuse FA abnormalities in SPG11 (França et al., 2012). However, this is not always the case, as FA and volume metrics are intrinsically different. For example, we have recently shown that in Down syndrome using TBSS analysis on FA reveals no differences between patients and controls, while DTBM detects highly significant differences in several brain regions (Pierpaoli et al., 2018). In another study of patients with Moebius syndrome, we found no significant differences in FA and TR between patients and controls, but DTBM detected atrophy in the posterior pons, including medial longitudinal fasciculus (Sadeghi et al., 2017, 2018). For SPG11 subjects, in contrast to previous DTI studies where diffuse white matter abnormalities were detected, DTBM was able to localize patterns of volume reduction within the white matter regions and to indicate a more severe and focal atrophy specifically affecting several long‐range pathways, including association, commissural, and projection pathways. Given that impaired axonal transport has been implicated in the pathogenesis of SPG11, this finding is consistent with the idea that long‐range pathways could be selectively affected in SPG11, because they are more vulnerable to impaired axonal transport.
One can also imagine other scenarios, where DTI measurements and morphometric measurements are affected differently. For example, consider a scenario in which the patient group is characterized by thinning of white matter pathways with preserved microstructure (i.e., fewer fibers, but with the remaining fibers structurally unaltered). In such a scenario, voxelwise analysis of FA should reveal no decrease in FA (although in practice we may find a reduction in FA, due to the contribution of partial volume contamination from the surrounding tissue). In this case, DTBM analysis would reveal the loss of volume due to a decrease in the number of fibers and provide a better understanding of the underlying change in the white matter structures. In another scenario, it may be that the structure of a fasciculus is altered in the patient group due to interstitial edema, but the overall size of the fasciculus is not changed—or even it may have increased. In this case, analysis of the LogJ maps may not reveal meaningful changes, while analysis of FA will show a remarkable reduction. In yet another scenario, involving gliosis accompanied by relatively minor loss of fiber in a brain region, voxelwise analysis of FA would be more informative as the architecture of the tissue may have changed without altering the overall size of the tissue.
4.2. Image processing factors and their influence on DTBM
One factor that could affect the result of analysis of LogJ maps is the choice of smoothing parameters during registration. DR‐TAMAS and ANTS use two sets of smoothing, one is the smoothing of the images and the other is the smoothing of the deformation fields. We use a similar strategy to smooth deformation fields, and we used the same value for the smoothing parameter across all methods when registering all subjects to the control template. In our experience, deformation field smoothing affects the LogJ maps the most and we used zero smoothing to preserve the location of atrophy and avoid blurring of tissue boundaries. Another parameter that could also affect the results is the step size used in the registration optimization algorithm. We also ensured that these parameters are consistent between the two programs. A small amount of smoothing can lead to a negative Jacobian, which is inconsistent with diffeomorphism. However, excessive smoothing can also blur the tissue boundaries. In this study, we decided to use limited image smoothing and no smoothing for deformation fields to preserve tissue boundaries and avoid blurring in the LogJ maps, however, this resulted in a negative Jacobian for a few voxels in the CC for SPG11 patients. In the future, development of the anisotropic smoothing kernel used during registration might remedy some of these issues.
It is worth noting that LogJ maps are also affected by the arrangement and sparsity of the fibers in the white matter bundle. For example, as is evident in Figure 1, a reduction in the CST volume is apparent in the LogJ maps of DTBM at the level of the medulla, but is not evident at the level of the pons. While taking this finding at face value may lead to the unlikely conclusion that the CST is differently impacted at different levels of the brainstem, a more plausible explanation is that the voxels containing the CST at the level of the medulla contain only CST fibers, whereas at the level of the pons they contain the CST as well as other pathways. In general, in regions containing multiple white matter pathways, the magnitude of the selective shrinkage of one of them will be diluted. For example, if pathway A represents 10% of the tissue in the voxel and its volume is reduced by 50%, the overall volume reduction in that voxel would be 5%, whereas if the pathway is the only white matter bundle in the voxel then the measured volume reduction would be 50%. Using multi‐fascicle registration methods such as the one proposed by Taquet et al. (2014) should improve DTBM in the crossing fiber regions.
Another important issue that could potentially affect the DTBM analysis is geometric distortion of the DTI data. We used registration of B0 images to undistorted T2W images to correct for distortion (Wu et al., 2008). However, one could use field maps or, if the data were acquired with multiple phase encoding directions, an algorithm such as DR‐BUDDI (Irfanoglu et al., 2015) can be used to correct for this distortion.
5. CONCLUSIONS
All TBM methods appear to detect white matter abnormalities, however, our findings suggest that DTBM is more informative and enables a better localization of white matter change within regions that appear homogenous in T1W images. Relative to FA‐TBM, DTBM shows more severe white matter atrophy and is able to detect enlargement of CSF spaces and atrophy of gray matter regions. As DR‐TAMAS takes the full diffusion tensor into account and uses deviatroic and Trace metrics when registering subjects to the control template, DTBM is able to detect differences even in regions with low anisotropy. DTBM combines the advantages of T1W‐TBM and FA‐TBM and provides a better delineation of atrophic regions. DTBM has the strength of T1W‐TBM in detecting atrophy of gray matter regions and enlargement of CSF spaces and the strength of FA‐TBM in localizing white matter atrophy that appears homogenous in T1W images with a greater specificity.
DTBM has potential for clinical applications in assessing brain morphology in a single subject or group comparison, as demonstrated, provided that normative data acquired with similar experimental parameters are available for the control population. A natural application of this method would be the analysis of volume changes of white matter pathways in brain development and aging, or using LogJ maps as an input to a classification algorithm. When atrophy or hypoplasia is severe, assessing group differences with deformation‐based analysis would be more appropriate conceptually than voxelwise analysis of DTI‐derived metrics, which would be biased by the presence of partial volume contamination.
Supporting information
Figure S1: Z‐scores of LogJ maps of less than −4 are overlaid on the control T1W template for each SPG11 subject using DTBM.
Figure S2: Z‐scores of LogJ maps of less than −4 are overlaid on the control T1W template for each SPG11 subject using T1W‐TBM. The brain is presented in radiological convention with the right hemisphere on the left of the image.
Figure S3: Z‐scores of LogJ maps of less than −4 are overlaid on the control T1W template for each SPG11 subject using FA‐TBM.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the NIH, including the National Institute of Biomedical Imaging and Bioengineering (NIBIB) and the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD). Support for this work included funding from the U01 HD079068‐01 grant, the Department of Defense in the Center for Neuroscience and Regenerative Medicine (CNRM) (HJF Award#: 30613610.0160855), and Congressionally Directed Medical Research Programs (CDMRP) (HJF Award#: W81XWH‐13‐2‐0019). Authors thank Dr. Henry Eden for his assistance with editing this manuscript.
Sadeghi N, Arrigoni F, D'Angelo MG, et al. Tensor‐based morphometry using scalar and directional information of diffusion tensor MRI data (DTBM): Application to hereditary spastic paraplegia. Hum Brain Mapp. 2018;39:4643–4651. 10.1002/hbm.24278
Funding information Center for Neuroscience and Regenerative Medicine (CNRM), Grant/Award Number: HJF Award#: 30613610.0160855; Congressionally Directed Medical Research Programs (CDMRP), Grant/Award Number: HJF Award#: W81XWH‐13‐2‐0019; National Institutes of Health (NIH), Grant/Award Number: U01 HD079068‐01
Footnotes
REFERENCES
- Ashburner, J. , & Friston, K. (2004). Morphometry In Frackowiak R. S. J., Friston K. J., Frith C., Dolan R., Friston K., Price C. J., … Penny W. D. (Eds.), Human brain function (2nd ed.). San Diego, California: Academic Press. [Google Scholar]
- Ashburner, J. , Hutton, C. , Frackowiak, R. , Johnsrude, I. , Price, C. , & Friston, K. (1998). Identifying global anatomical differences: Deformation‐based morphometry. Human Brain Mapping, 6(56), 348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants, B. B. , Epstein, C. L. , Grossman, M. , & Gee, J. C. (2008). Symmetric diffeomorphic image registration with cross‐correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis, 12(1), 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants, B. B. , Tustison, N. J. , Song, G. , Cook, P. A. , Klein, A. , & Gee, J. C. (2011). A reproducible evaluation of ANTs similarity metric performance in brain image registration. NeuroImage, 54(3), 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants, B. B. , Yushkevich, P. , Pluta, J. , Minkoff, D. , Korczykowski, M. , Detre, J. , & Gee, J. C. (2010). The optimal template effect in hippocampus studies of diseased populations. NeuroImage, 49(3), 2457–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser, P. , & Pierpaoli, C. (1996). Microstructural and physiological features of tissues elucidated by quantitative‐diffusion‐tensor MRI. Journal of Magnetic Resonance, Series B, 111(3), 209–219. [DOI] [PubMed] [Google Scholar]
- Basser, P. J. , Mattiello, J. , & LeBihan, D. (1994). Estimation of the effective self‐diffusion tensor from the NMR spin echo. Journal of Magnetic Resonance, Series B, 103(3), 247–254. [DOI] [PubMed] [Google Scholar]
- Cao, L. , Rong, T.‐Y. , Huang, X.‐J. , Fang, R. , Wu, Z.‐Y. , Tang, H.‐D. , & Chen, S.‐D. (2013). Novel SPG11 mutations in Chinese families with hereditary spastic paraplegia with thin corpus callosum. Parkinsonism & Related Disorders, 19(3), 367–370. [DOI] [PubMed] [Google Scholar]
- Catani, M. , & de Schotten, M. T. (2012). Atlas of human brain connections (p. 11). New York: Oxford University Press. [Google Scholar]
- Chang, L.‐C. , Jones, D. K. , & Pierpaoli, C. (2005). Restore: Robust estimation of tensors by outlier rejection. Magnetic Resonance in Medicine, 53(5), 1088–1095. [DOI] [PubMed] [Google Scholar]
- Chang, L. C. , Walker, L. , & Pierpaoli, C. (2012). Informed RESTORE: A method for robust estimation of diffusion tensor from low redundancy datasets in the presence of physiological noise artifacts. Magnetic Resonance in Medicine, 68(5), 1654–1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Q. , Lui, S. , Wang, J.‐G. , Ou‐Yang, L. , Zhou, D. , Burgunder, J.‐M. , … Shang, H.‐F. (2008). Diffusion tensor imaging of two unrelated Chinese men with hereditary spastic paraplegia associated with thin corpus callosum. Neuroscience Letters, 441(1), 21–24. [DOI] [PubMed] [Google Scholar]
- Chiang, M.‐C. , Dutton, R. A. , Hayashi, K. M. , Lopez, O. L. , Aizenstein, H. J. , Toga, A. W. , … Thompson, P. M. (2007). 3D pattern of brain atrophy in HIV/AIDS visualized using tensor‐based morphometry. NeuroImage, 34(1), 44–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen, J. (1988). Statistical power analysis for the behavioral sciences. 2nd ed. New York, NY: Academic Press/Elsevier. [Google Scholar]
- Davatzikos, C. , Vaillant, M. , Resnick, S. M. , Prince, J. L. , Letovsky, S. , & Bryan, R. N. (1996). A computerized approach for morphological analysis of the corpus callosum. Journal of Computer Assisted Tomography, 20(1), 88–97. [DOI] [PubMed] [Google Scholar]
- Deluca, G. , Ebers, G. , & Esiri, M. (2004). The extent of axonal loss in the long tracts in hereditary spastic paraplegia. Neuropathology and Applied Neurobiology, 30(6), 576–584. [DOI] [PubMed] [Google Scholar]
- Fink, J. K. (2013). Hereditary spastic paraplegia: Clinico‐pathologic features and emerging molecular mechanisms. Acta Neuropathologica, 126(3), 307–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- França, M. C. , Yasuda, C. L. , Pereira, F. R. , D'Abreu, A. , Lopes‐Ramos, C. M. , Rosa, M. V. , … Lopes‐Cendes, I. (2012). White and grey matter abnormalities in patients with SPG11 mutations. Journal of Neurology, Neurosurgery & Psychiatry, 83(8), 828–833. [DOI] [PubMed] [Google Scholar]
- Garaci, F. , Toschi, N. , Lanzafame, S. , Meschini, A. , Bertini, E. , Simonetti, G. , … Floris, R. (2014). Diffusion tensor imaging in SPG11‐and SPG4‐linked hereditary spastic paraplegia. International Journal of Neuroscience, 124(4), 261–270. [DOI] [PubMed] [Google Scholar]
- Hedges, L. V. (1981). Distribution theory for glass's estimator of effect size and related estimators. Journal of Educational Statistics, 6(2), 107–128. [Google Scholar]
- Hua, X. , Leow, A. D. , Parikshak, N. , Lee, S. , Chiang, M.‐C. , Toga, A. W. , … Thompson, P. M. (2008). Tensor‐based morphometry as a neuroimaging biomarker for Alzheimer's disease: An MRI study of 676 AD, MCI, and normal subjects. NeuroImage, 43(3), 458–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu, M. O. , Modi, P. , Nayak, A. , Hutchinson, E. B. , Sarlls, J. , & Pierpaoli, C. (2015). DR‐BUDDI (diffeomorphic registration for blip‐up blip‐down diffusion imaging) method for correcting echo planar imaging distortions. NeuroImage, 106, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu, M. O. , Nayak, A. , Jenkins, J. , Hutchinson, E. , Sadeghi, N. , Thomas, C. , & Pierpaoli, C. (2016). DR‐TAMAS: Diffeomorphic registration for tensor accurate alignment of anatomical structures. NeuroImage, 132, 439–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu, M.O. , Nayak A., Jenkins J., Pierpaoli C. (2017). TORTOISE V3: improvements and new features of the NIH diffusion MRI processing pipeline. ISMRM 25th Annual Meeting, Honolulu, HI.
- Irfanoglu, M.O. , Sadeghi, N. , Nayak, A. , Pierpaoli, C. (2017). Strategies for building a morphologically faithful average brain template from population diffusion MRI data. ISMRM 25th Annual Meeting, Honolulu, HI.
- Kipps, C. , Duggins, A. , Mahant, N. , Gomes, L. , Ashburner, J. , & McCusker, E. (2005). Progression of structural neuropathology in preclinical Huntington's disease: A tensor based morphometry study. Journal of Neurology, Neurosurgery & Psychiatry, 76(5), 650–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, A. , Andersson, J. , Ardekani, B. A. , Ashburner, J. , Avants, B. , Chiang, M.‐C. , … Parsey, R. V. (2009). Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage, 46(3), 786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier‐Hein, K. H. , Neher, P. F. , Houde, J. C. , Côté, M. A. , Garyfallidis, E. , Zhong, J. , … Reddick, W. E. (2017). The challenge of mapping the human connectome based on diffusion tractography. Nature Communications, 8(1), 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott, C. , White, K. , Bushby, K. , & Shaw, P. (2000). Hereditary spastic paraparesis: A review of new developments. Journal of Neurology, Neurosurgery & Psychiatry, 69(2), 150–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, S. , & van Zijl, P. (2002). Fiber tracking: Principles and strategies—a technical review. NMR in Biomedicine, 15(7–8), 468–480. [DOI] [PubMed] [Google Scholar]
- Murphy, K. , Van Ginneken, B. , Reinhardt, J. M. , Kabus, S. , Ding, K. , Deng, X. , … Pluim, J. P. W. (2011). Evaluation of registration methods on thoracic CT: The empire10 challenge. IEEE Transactions on Medical Imaging, 30(11), 1901–1920. [DOI] [PubMed] [Google Scholar]
- Oishi, K. , Akhter, K. , Mielke, M. , Ceritoglu, C. , Zhang, J. , Jiang, H. , … Mori, S. (2011). Multi‐modal MRI analysis with disease‐specific spatial filtering: Initial testing to predict mild cognitive impairment patients who convert to Alzheimer's disease. Frontiers in Neurology, 2, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani, E. , Horsfield, M. A. , Rocca, M. A. , & Filippi, M. (2007). Assessing atrophy of the major white matter fiber bundles of the brain from diffusion tensor MRI data. Magnetic Resonance in Medicine, 58(3), 527–534. [DOI] [PubMed] [Google Scholar]
- Pajevic, S. , & Pierpaoli, C. (1999). Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: Application to white matter fiber tract mapping in the human brain. Magnetic Resonance in Medicine, 42(3), 526–540. [PubMed] [Google Scholar]
- Pérez‐Brangulí, F. , Mishra, H. K. , Prots, I. , Havlicek, S. , Kohl, Z. , Saul, D. , … Sock, E. (2014). Dysfunction of spatacsin leads to axonal pathology in SPG11‐linked hereditary spastic paraplegia. Human Molecular Genetics, 23(18), 4859–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli, C. , Nayak, A. , Irfanoglu, M. O. , Sadeghi, N. , & Raitano‐Lee, N. (2018). Brain morphometry using diffusion MRI data (DTBM) reveals abnormalities in Down Syndrome that are not detected by conventional DTI analysis Paris, France: ISMRM. [Google Scholar]
- Pierpaoli, C. , Jezzard, P. , Basser, P. J. , Barnett, A. , & Di Chiro, G. (1996). Diffusion tensor MR imaging of the human brain. Radiology, 201(3), 637–648. [DOI] [PubMed] [Google Scholar]
- Pierpaoli, C ., Walker, L ., Irfanoglu, M ., Barnett, A ., Chang, L.‐C ., Koay, C ., …, Wu, M . (2010). TORTOISE: An integrated software package for processing of diffusion MRI data. ISMRM 19th Annual Meeting, Stockholm, Sweden.
- Rohde, G. K. , Barnett, A. S. , Basser, P. J. , Marenco, S. , & Pierpaoli, C. (2004). Comprehensive approach for correction of motion and distortion in diffusion‐weighted MRI. Magnetic Resonance in Medicine, 51(1), 103–114. [DOI] [PubMed] [Google Scholar]
- Sadeghi, N. , Manoli, I. , Hutchinson, E. , Van Ryzin, C. , Thomas, C. , Irfanoglu, M.O. , …, The Moebius Collaborative Research Group (2017). Brain morphometry driven by DTI data in Moebius syndrome and hereditary congenital facial paresis. Annual Meeting of the Organization for Human Brain Mapping (OHBM), Vancouver, Canada.
- Sadeghi, N. , Manoli, I. , Wood, T. , Collins, F. S. , Jabs, E. W. , Engle, E. C. , & Pierpaoli, C. (2018). Features extracted from diffusion‐driven tensor based morphometry can serve as a specific imaging marker for Moebius syndrome. Paris, France: ISMRM. [Google Scholar]
- Salinas, S. , Proukakis, C. , Crosby, A. , & Warner, T. T. (2008). Hereditary spastic paraplegia: Clinical features and pathogenetic mechanisms. The Lancet Neurology, 7(12), 1127–1138. [DOI] [PubMed] [Google Scholar]
- Stevanin, G. , Santorelli, F. M. , Azzedine, H. , Coutinho, P. , Chomilier, J. , Denora, P. S. , … Brice, A. (2007). Mutations in SPG11, encoding spatacsin, are a major cause of spastic paraplegia with thin corpus callosum. Nature Genetics, 39(3), 366–372. [DOI] [PubMed] [Google Scholar]
- Studholme, C. (2008). Dense feature deformation morphometry: Incorporating DTI data into conventional MRI morphometry. Medical Image Analysis, 12(6), 742–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taquet, M. , Scherrer, B. , Commowick, O. , Peters, J. M. , Sahin, M. , Macq, B. , & Warfield, S. K. (2014). A mathematical framework for the registration and analysis of multi‐fascicle models for population studies of the brain microstructure. IEEE Transactions on Medical Imaging, 33(2), 504–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C. , Frank, Q. Y. , Irfanoglu, M. O. , Modi, P. , Saleem, K. S. , Leopold, D. A. , & Pierpaoli, C. (2014). Anatomical accuracy of brain connections derived from diffusion MRI tractography is inherently limited. Proceedings of the National Academy of Sciences, 111(46), 16574–16579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, P. , Gledd, J. , Woods, R. , MacDonald, D. , Evans, A. , & Toga, A. (2000). Growth patterns in the developing brain detected by using continuum mechanical tensor maps. Nature, 404(6774), 190–193. [DOI] [PubMed] [Google Scholar]
- Verma, R. , Mori, S. , Shen, D. , Yarowsky, P. , Zhang, J. , & Davatzikos, C. (2005). Spatiotemporal maturation patterns of murine brain quantified by diffusion tensor MRI and deformation‐based morphometry. Proceedings of the National Academy of Sciences of the United States of America, 102(19), 6978–6983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Benner, T. , Sorensen, A. , & Wedeen, V. (2007). Diffusion toolkit: A software package for diffusion imaging data processing and tractography. Proceedings of the International Society for Magnetic Resonance in Medicine, 15, 3720. [Google Scholar]
- Wharton, S. B. , McDermott, C. J. , Grierson, A. J. , Wood, J. D. , Gelsthorpe, C. , Ince, P. G. , & Shaw, P. J. (2003). The cellular and molecular pathology of the motor system in hereditary spastic paraparesis due to mutation of the spastin gene. Journal of Neuropathology & Experimental Neurology, 62(11), 1166–1177. [DOI] [PubMed] [Google Scholar]
- White, K. , Ince, P. , Lusher, M. , Lindsey, J. , Cookson, M. , Bashir, R. , … Bushby, K. (2000). Clinical and pathologic findings in hereditary spastic paraparesis with spastin mutation. Neurology, 55(1), 89–94. [DOI] [PubMed] [Google Scholar]
- Winkler, A. M. , Ridgway, G. R. , Webster, M. A. , Smith, S. M. , & Nichols, T. E. (2014). Permutation inference for the general linear model. NeuroImage, 92, 381–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, M. , Chang, L. C. , Walker, L. , Lemaitre, H. , Barnett, A. S. , Marenco, S. , & Pierpaoli, C. (2008). Comparison of EPI distortion correction methods in diffusion tensor MRI using a novel framework. Medical Image Computing and Computer‐Assisted Intervention, 11(Pt 2), 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, H. , Avants, B. B. , Yushkevich, P. A. , Woo, J. H. , Wang, S. , McCluskey, L. F. , … Gee, J. C. (2007). High‐dimensional spatial normalization of diffusion tensor images improves the detection of white matter differences: An example study using amyotrophic lateral sclerosis. IEEE Transactions on Medical Imaging, 26(11), 1585–1597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Z‐scores of LogJ maps of less than −4 are overlaid on the control T1W template for each SPG11 subject using DTBM.
Figure S2: Z‐scores of LogJ maps of less than −4 are overlaid on the control T1W template for each SPG11 subject using T1W‐TBM. The brain is presented in radiological convention with the right hemisphere on the left of the image.
Figure S3: Z‐scores of LogJ maps of less than −4 are overlaid on the control T1W template for each SPG11 subject using FA‐TBM.


