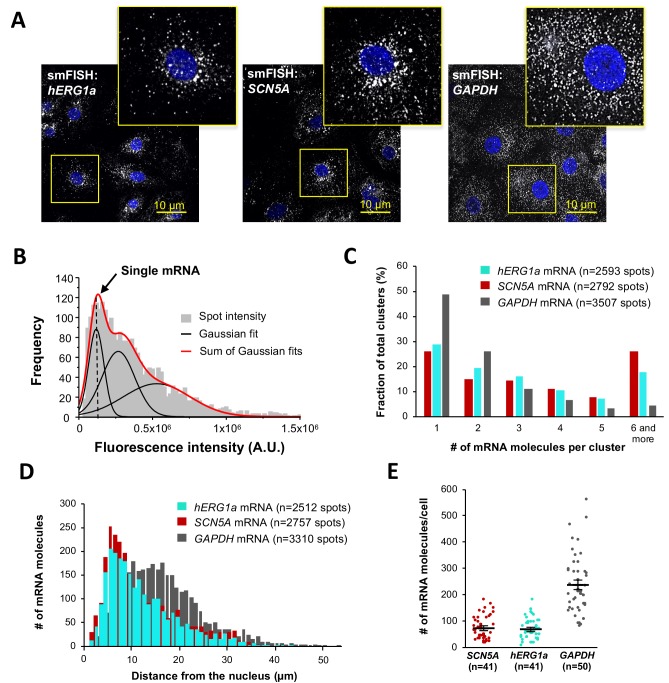
Figure 2. Quantitative description of single hERG1a and SCN5A transcripts and their distribution in iPSC-CMs.
(A) Representative confocal images and enlargement (outlined in yellow) of iPSC-CMs subjected to the smFISH protocol. (B) By fitting the intensity histogram of smFISH signals (n = 2611 spots) to the sum of Gaussian functions (red line), the typical intensity corresponding to a single mRNA molecule (vertical dashed line) was extracted. (C) The distribution of the number of mRNA molecules associated in clusters for each transcript evaluated by smFISH. (D) Histogram showing the cytoplasmic distribution of mRNA signals with distance from the nucleus. (E) The number of mRNAs detected per cell was plotted for SCN5A, hERG1a and GAPDH (lines represent mean ±SE).