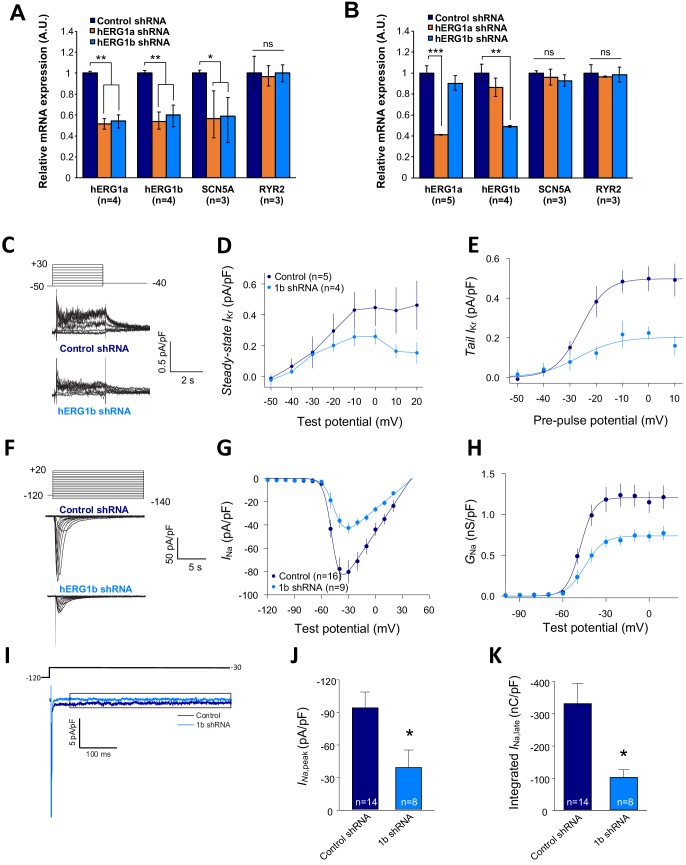
Figure 7. Co-knockdown of IKr and INa by hERG transcript-specific shRNA.
(A) Effects of hERG1a or hERG1b silencing on channel mRNA expression levels detected by RT-qPCR (mean ±95% CI) in iPSC-CMs. A non-targeting shRNA (scrambled shRNA) is used as a control. (B) Effects of specific hERG1a or hERG1b silencing on ion channel mRNAs expressed alone in HEK293 cells. (C) Representative family of traces show IKr in presence of control (upper) or hERG1b shRNA (lower). (D) Summary of steady-state current density vs. test potential shows effect of hERG1b shRNA (mean ±SE). (E) Effects of 1b shRNA on peak tail current vs. pre-pulse potential (mean ±SE). (F) Representative family of traces recorded from iPSC-CMs showing effects of hERG1b-specific shRNA compared to control shRNA on peak INa. (G) Summary current-voltage plot of peak INa vs. test potential (mean ±SE). (H) Summary conductance (G)-voltage plot based on data from G (mean ±SE). (I) Late sodium current representative trace in control and 1b shRNA-transfected cells, measured by applying a single pulse protocol of 800 ms. (J) Summary statistics of peak INa showed a decrease upon transfection with hERG1b shRNA (mean ±SE). (K) Late INa measured as the integral from 50 to 800 ms from the beginning of the pulse showed a decrease upon transfection with hERG1b shRNA (mean ±SE).