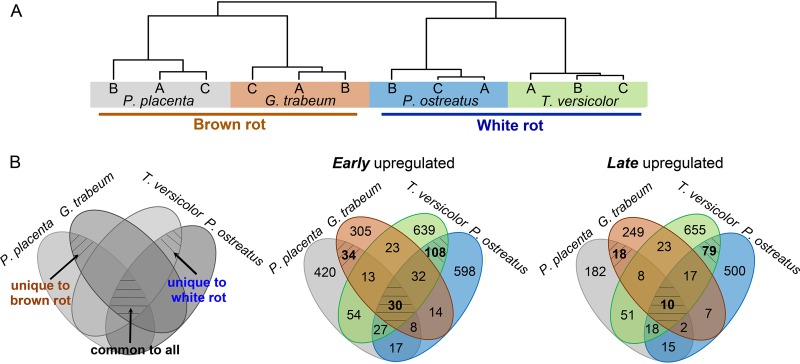
FIG 2.
Differential gene expression as a function of species and wood decay stage. (A) The similarity of global temporal gene regulation among species clearly indicated two major clusters distinguished by nutritional modes of brown rot and white rot. The temporal gene expression trends of 2,093 orthologous DEGs were used to calculate the correlations among fungal samples by Spearman’s method. “A,” “B,” and “C” represent triple bioreplicates of RNA-seq data in each species. (B) Unique and common differentially expressed genes (DEGs) as a function of species and wood decay stage. The “decay-stage-dependent” genes, i.e., genes upregulated “early” or “late,” were compared among four species with the orthologous genes. The same MCL ID was assigned to genes from one ortho group, while species-specific ID was used for that without orthologues in other species. “Late” DEGs were combined from 15- to 20-mm and 30- to 35-mm sections. The brown rot fungi are P. placenta and G. trabeum, and the white rot fungi are T. versicolor and P. ostreatus.