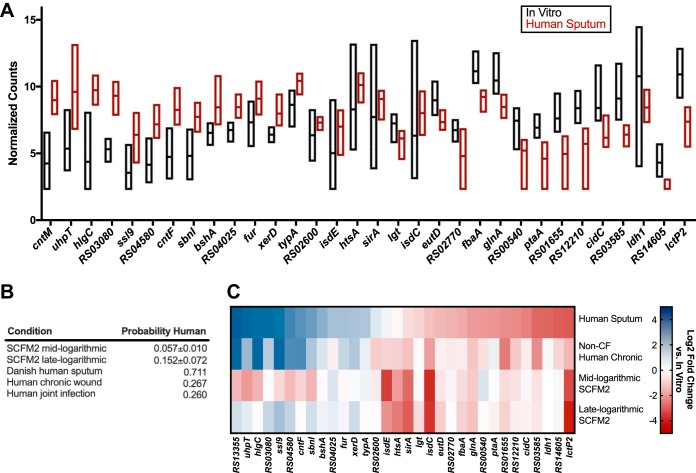
FIG 3.
A subset of 32 genes is sufficient to distinguish S. aureus transcriptomes from human CF lung infection and those grown in vitro. (A) The x axis lists the 32 genes used to build our SVM model. VST-normalized read counts are plotted on the y axis. Black bars are samples grown in vitro (n = 22), and red bars are samples from human CF sputum (n = 9). Bars indicate the range of counts with a line that indicates the mean within the group. (B) Table showing the probability of the transcriptome from each condition to be from a human infection when classified by our SVM model. The standard deviation of the probability for either 4 replicates (mid-logarithmic) or 5 replicates (late logarithmic) is shown for the SCFM2 conditions. (C) Heat map showing the log2 fold change of S. aureus transcriptomes in human sputum, non-CF human chronic infection, mid-logarithmic growth in SCFM2, or late-logarithmic growth in SCFM2 compared to in vitro growth for the 32-gene transcriptomic signature used in the SVM model.