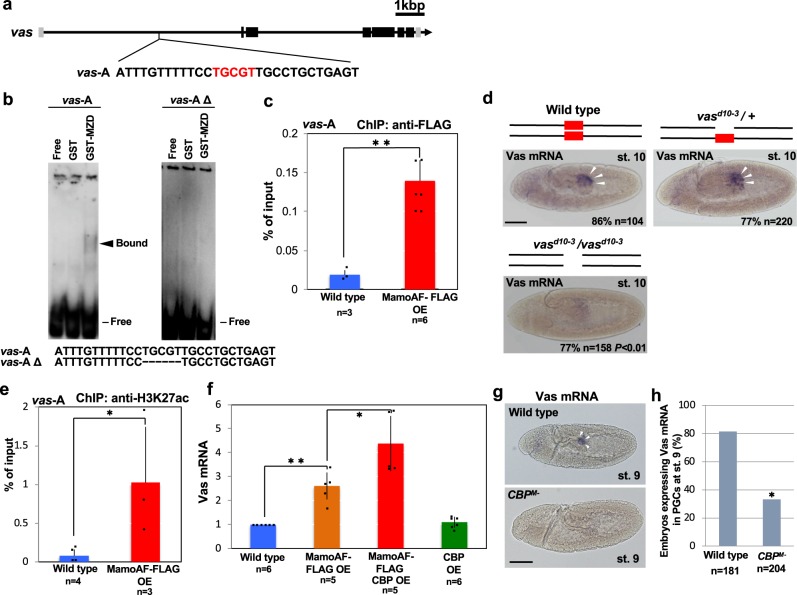
Fig. 2.
MamoAF directly binds vas and epigenetically activates vas together with CBP. a vas-A element, which contains a Mamo-binding consensus sequence (red), in the first intron of vas. b EMSA analyses of Zn-finger domains of Mamo protein (MZD) using vas-A and vas-AΔ probes. vas-AΔ probe lacks a Mamo-binding consensus sequence. c ChIP-qPCR assays were performed with MamoAF-OE embryos and FLAG antibody. The precipitated DNA was detected by qPCR with primers targeting to vas-A. Error bars are SD of the mean. **P < 0.01 (two-tailed Student’s t test). d Vas mRNA in situ hybridisation of wild type, vasd10-3 / + and vasd10-3 / vasd10-3 embryos at stage 10. The Mamo-binding consensus sequence (red box) is deleted from vas-A element in vasd10-3 / vasd10-3embryo. Scale bar: 50 μm. e ChIP-qPCR assays were performed with MamoAF-OE embryos and H3K27ac antibody. The precipitated DNA was detected by qPCR with primers targeting to vas-A. Error bars are SD of the mean. *P < 0.05 (one-tailed Student’s t test). f qRT-PCR analyses of Vas mRNA in wild type, MamoAF, CBP and both CBP and MamoAF overexpressed embryos. Error bars are SD of the mean. *P < 0.05, **P < 0.01 (two-tailed Student’s t test). g Vas mRNA in situ hybridisation of wild type and CBPM- embryo derived from nej0.3 mutant germline clones at stage 9. White arrowheads indicate PGCs expressing Vas mRNA. Scale bar: 50 μm. h Percentages of embryos expressing Vas mRNA in wild type and CBPM- embryos at stage 9. *P < 0.01 (Fisher’s exact test)