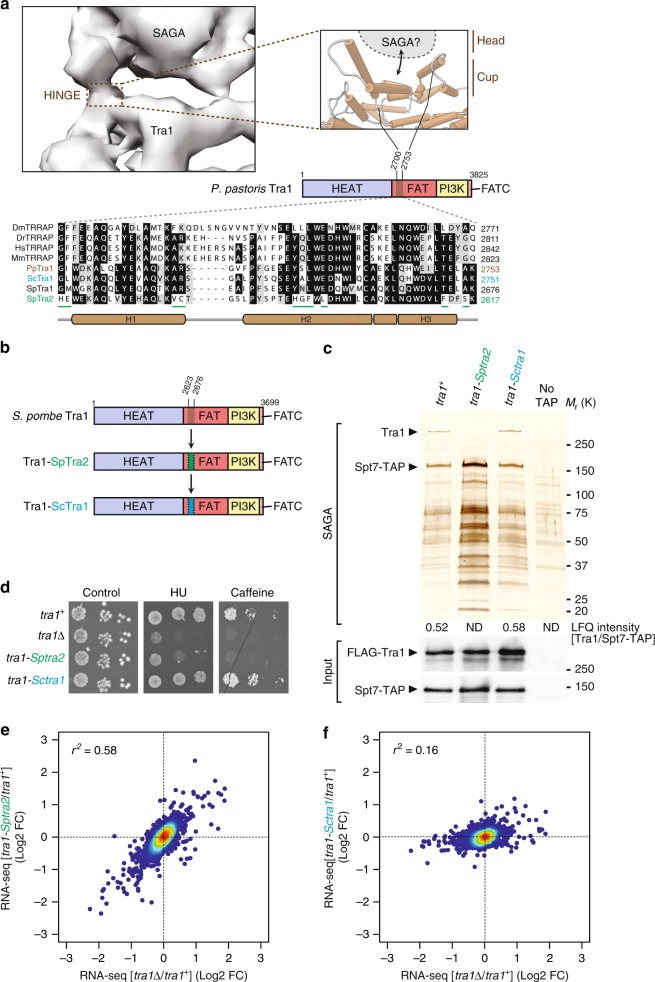
Fig. 5.
Mechanism of Tra1 specific interaction with SAGA. a Close-up view of the putative region of Tra1 that contacts the rest of SAGA, which constitutes the flexible hinge in the structure of P. pastoris SAGA (EMD: 3804). Cartoon cylinders represent α-helices in P. Pastoris Tra1 structure (PDB: 5OEJ). This domain is located near the start of the FAT domain and corresponds to residues 2700–2753 (brown-coloured box). A homologous region, defined as the Cup SAGA Interacting (CSI), was identified in S. pombe Tra1 (residues 2623–2676) from multiple alignments of Tra1 orthologs, shown at the bottom. Residues that appear unique to S. pombe Tra2 are underlined (green). b Schematic illustration of the hybrid mutant alleles of S. pombe tra1+ that were constructed. Residues 2623–2676 from S. pombe Tra1 were swapped with the homologous region from either S. pombe Tra2 (green, residues 2564–2617), to create the tra1-Sptra2 allele, or S. cerevisiae Tra1 (blue, residues 2698–2751), to create the tra1-Sctra1 allele. c Silver staining of SAGA complexes purified from WT, tra1-Sptra2 and tra1-Sctra1 strains (see b), using Spt7 as the bait. A non-tagged strain (no TAP) was used as a control for background. Numbers at the bottom of the gel represent LFQ intensity ratios of Tra1 to the bait, Spt7, from LC-MS/MS analyses of purified SAGA complexes (ND: not detected or <1% of WT). Below are anti-FLAG and anti-HA western blotting of FLAG-Tra1 and Spt7-TAP in a fraction of the input used for TAP. Data are representative of five independent experiments. d–f HU sensitivity (d) and gene expression changes (e, f) of tra1-Sptra2 and tra1-Sctra1 strains, as compared with tra1Δ mutants. d Tenfold serial dilutions of exponentially growing cells of the indicated genotypes were spotted on rich medium (control), medium supplemented with 10 mM HU, or 15 mM caffeine, and incubated at 32 °C. e, f Density scatter plots from RNA-seq data comparing tra1Δ mutants (x-axis) with either tra1-Sptra2 mutants (y-axis in e) or tra1-Sctra1 mutants (y-axis in f), relative to isogenic WT controls (n = 3 independent biological samples). Statistical significance and correlation were analysed by computing the Pearson coefficient of determination (r2 = 0.58 for tra1-Sptra2 vs. tra1Δ and r2 = 0.16 for tra1-Sctra1 vs. tra1Δ; P < 0.0001). Source data are provided as a Source Data file