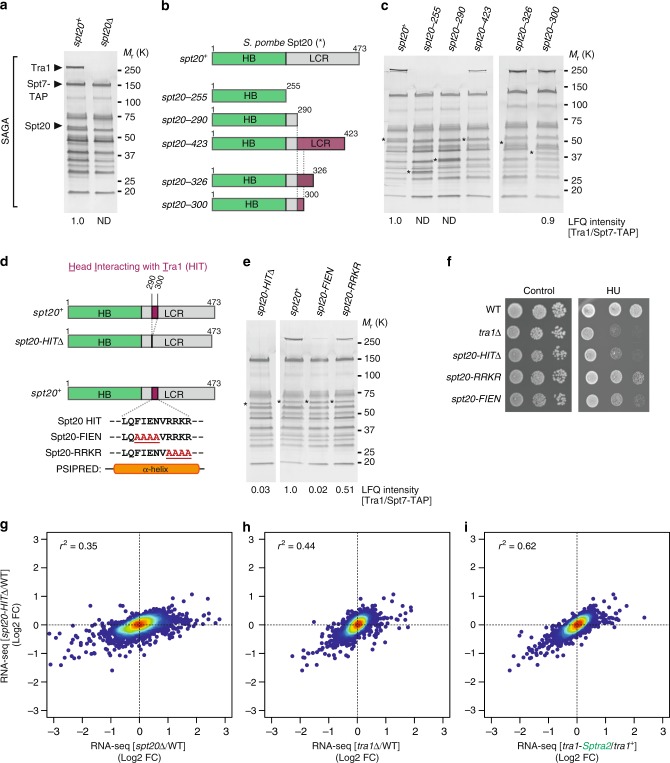
Fig. 6.
Spt20 anchors Tra1 into the SAGA complex. a Silver staining of SAGA complexes purified from WT and spt20Δ strains, using Spt7 as the bait. A band corresponding to Spt20 disappears in spt20Δ mutants. b Schematic illustration of the different Spt20 truncation mutants constructed to identify the Head Interacting with Tra1 (HIT) region of Spt20 from S. pombe. Distinct colours depict Spt20 domains, defined as Homology Boxes (HB) and a Low Complexity Region (LCR). Each mutant is named after to the last residue present in the truncation mutant, which shortens the LCR to various extent, as illustrated. c Silver staining of SAGA complexes purified from WT and spt20 truncation mutants, using Spt7 as the bait. Numbers at the bottom of the gel represent LFQ intensity ratios of Tra1 to Spt7, from LC-MS/MS analyses of purified SAGA complexes (ND: not detected or <1% of WT). Purple colouring depicts the region of Spt20 required for Tra1-SAGA interaction as inferred from each truncation mutant. Asterisks indicate the position of the WT and mutant Spt20 proteins in purified SAGA. d Schematic illustration of the deletion or point mutations within Spt20 HIT region, narrowed down to residues 290–300 (purple-coloured box) and predicted to fold into a α-helix, using PSI-blast based secondary structure PREDiction (PSIPRED70). e Silver staining of SAGA complexes purified from WT, spt20-HITΔ, spt20-FIEN and spt20-RRKR mutants, using Spt7 as the bait. Numbers at the bottom of the gel represent LFQ intensity ratios of Tra1 to Spt7, from LC-MS/MS analyses of purified SAGA complexes. Values for each mutant are expressed as percentage of WT SAGA. Asterisks indicate the position of the WT and mutant Spt20 proteins in purified SAGA. f HU sensitivity of spt20-HITΔ, spt20-FIEN and spt20-RRKR mutants, as compared with tra1Δ mutant strains. Tenfold serial dilutions of exponentially growing cells of the indicated genotypes were spotted either on rich medium (control) or medium supplemented with 5 mM HU and incubated for 3 days at 32 °C. g–i Density scatter plots plots from RNA-seq data comparing spt20-HITΔ mutants (y-axis) with either spt20Δ mutants (x-axis in g), tra1Δ mutants (x-axis in h), or tra1-Sptra2 mutants (x-axis in i), relative to isogenic WT controls (n = 3 independent biological samples). Statistical significance and correlation were analysed by computing the Pearson coefficient of determination (r2 = 0.35 for spt20-HITΔ vs. spt20Δ; r2 = 0.44 for spt20-HITΔ vs. tra1Δ; r2 = 0.62 for spt20-HITΔ vs. tra1-Sptra2; P < 0.0001). Source data are provided as a Source Data file