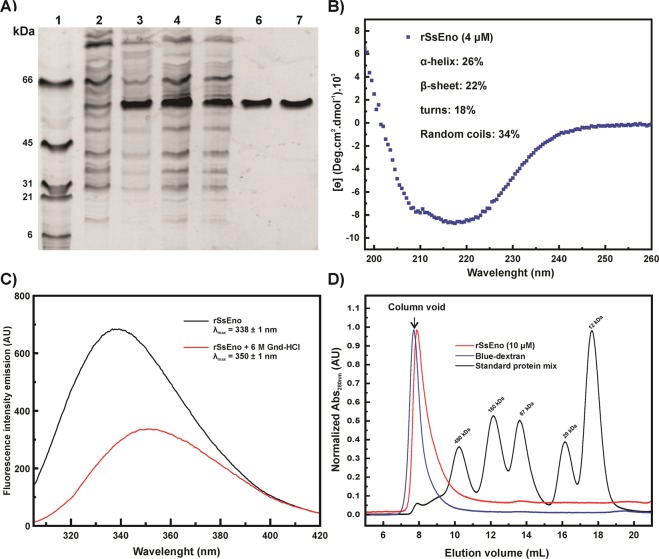
Figure 1.
SDS-PAGE and structure analysis of rSsEno expressed in E. coli BL21. The recombinant plasmid pET28a::SsEno-transformed E. coli BL21 cells were induced in the presence of 0,2 mM IPTG for 4 h at 30 °C. The cells were lysed by sonication, and the supernatant containing the recombinant protein was purified by affinity and preparative SEC, respectively. All the samples were analyzed by SDS-PAGE 12%, and the protein was stained with Coomassie Blue R250 in the gel. (A) Expression and purification of rSsEno. Molecular mass markers in kDa (lane 1), non-IPTG-induced pET28a::SsEno-transformed E. coli BL21 cells lysate (lane 2), IPTG-induced pET28a::SsEno-transformed E. coli BL21 cells lysate (lane 3); supernatant of lysed IPTG-induced pET28a::SsEno-transformedE. coli BL21 cells (lane 4), supernatant of lysed cells filtered through Hydrophlic Durapore Membrane, 0.45 µm cutoff, 47 mm diameter (lane 5), rSsEno purified by Ni2+ affinity chromatography (lane 6) and by preparative SEC (lane 7). (B) The CD spectrum shows that rSsEno was obtained mainly with a secondary structure composed by α-helices and β-sheets. (C) Intrinsic emission fluorescence spectra for enolase in the folded (black curve) and unfolded (red curve) states induced by 6 M Gnd-HCl. (D) Analytical SEC performed for rSsEno (red line). The MW standard protein mix elution pattern is represented by the black line: 1) Apoferritin (480 kDa); 2) γ-Globulin (160 kDa); 3) BSA (67 kDa); 4) carbonic anhydrase (29 kDa); 5) Cytochrome C (12 kDa). The column void is identified by blue dextran (blue line).