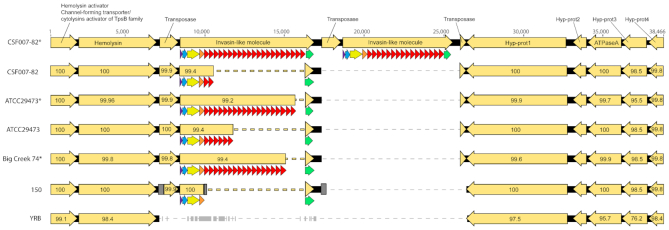
Figure 1.
DNA alignment of a ∼39 kb-long DNA region containing the yrIlm gene and flanking CDS in Y. ruckeri genomes deposited in GenBank. Each CDS is indicated by a yellow arrow, with the percentage of sequence identity to CSF007-82 reported inside the arrow. yrIlm consists of an array of tandemly repeated, identical Ig-like domains (in red) and in addition of Ig-like domains of lower pairwise sequence similarity (in orange). It is usually capped by a C-type lectin domain (CTLD, in green). The dashed lines indicate gaps in the DNA alignment. In strain 150 the grey box indicates a contig break in the assembly. The asterisk (*) indicates assemblies generated through PacBio SMRT sequencing. Note that the other assemblies have significant lower repeat numbers, suggesting that the repeats were not found using short-read sequencing technologies. Modified from Wrobel,A., Ottoni,C., Leo,J.C., Gulla,S. and Linke,D. (2018) The repeat structure of two paralogous genes, Yersinia ruckeri invasin (yrInv) and a ‘Y. ruckeri invasin-like molecule’, (yrIlm) sheds light on the evolution of adhesive capacities of a fish pathogen. Journal of Structural Biology, 201, 171–183, with permission from Elsevier.