Figure 1.
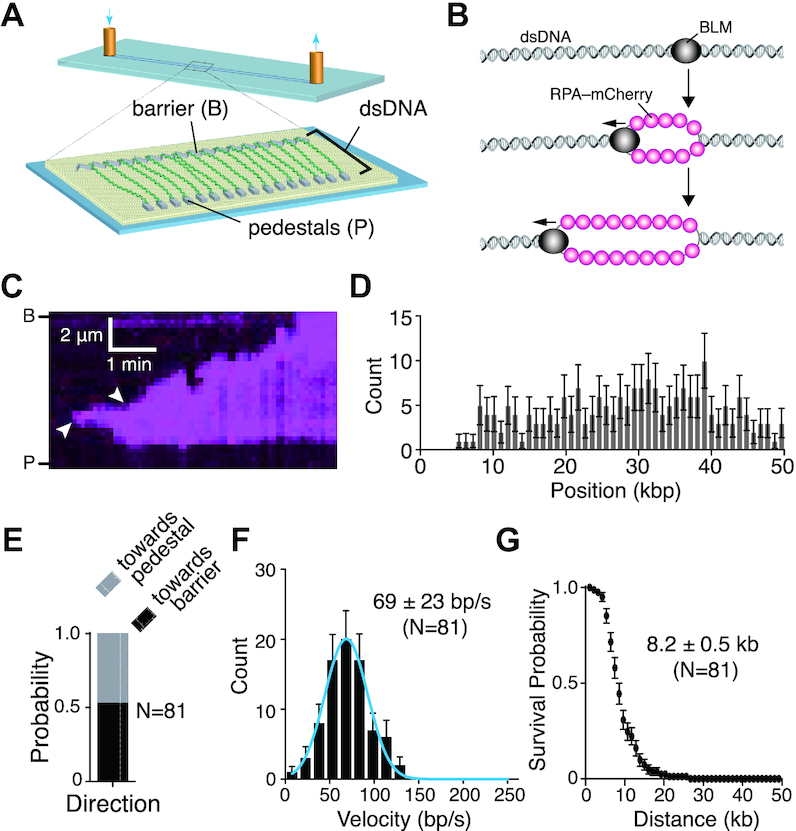
BLM is a fast and highly processive dsDNA helicase. (A) Schematic illustration of the double-tethered DNA curtains assay. (B) Schematic showing experimental rational for the detection of dsDNA unwinding activity for unlabeled BLM as revealed by the binding of RPA-mCherry to the resulting ssDNA products. (C) Kymograph showing BLM (unlabeled) unwinding dsDNA (unlabeled) as revealed by the binding of RPA-mCherry (magenta); note that buffer flow was OFF during data collection. Arrowheads indicate the sites where BLM initiated dsDNA unwinding based upon the appearance of RPA-mCherry. Reactions contained 0.2 nM unlabeled BLM and 1 nM RPA-mCherry. (D) Distribution of sites where BLM initiated dsDNA unwinding; error bars represent 95% confidence intervals calculated from bootstrap analysis. (E) Quantification of BLM translocation direction in reactions with 1 nM RPA-mCherry. ‘Towards pedestal’ indicates BLM movement in the direction from the barrier to the pedestal, and ‘towards barrier’ indicates movement in the opposite direction. (F) Velocity distribution of BLM unwinding rates in reactions with 1 nM RPA-mCherry on double-tethered dsDNA. The solid blue line represents a Gaussian fit to the data. Error bars represent 95% confidence intervals calculated from bootstrap analysis. (G) Survival probability plot of BLM translocation with 1 nM RPA-mCherry on double-tethered dsDNA. Error bars represent 95% confidence intervals calculated from bootstrap analysis.
