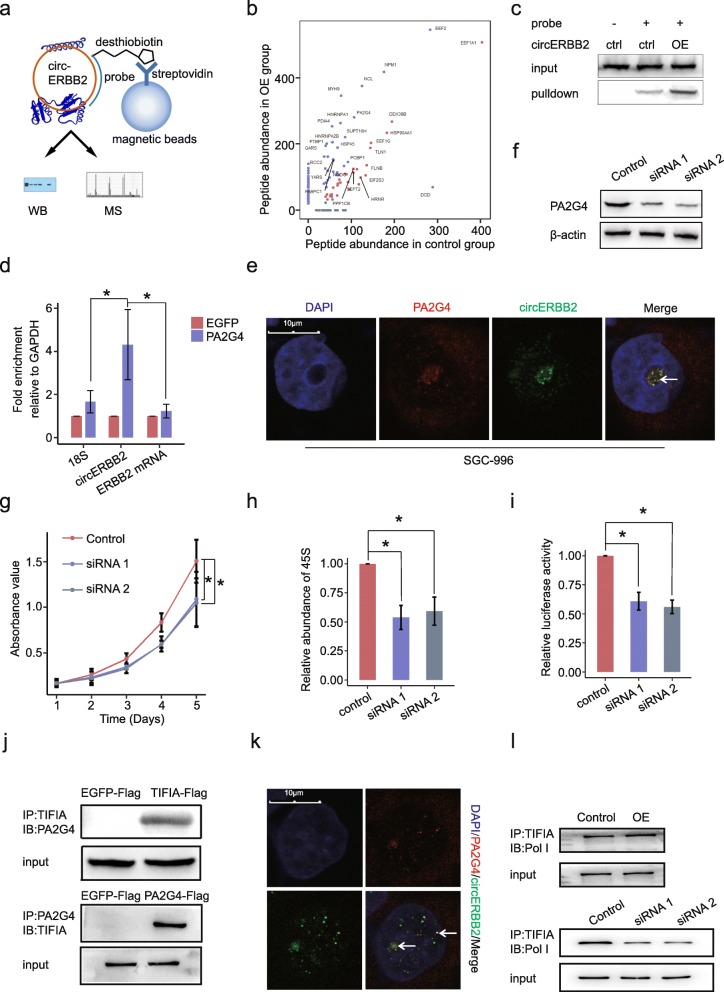
Fig. 5.
circERBB2 interacts with PA2G4. a Schematic plot of circRNA pulldown assay with a desthiobiotin-labeled DNA probe. b Potential circERBB2-associated proteins identified by mass spectrum. Proteins in purple, which had significant higher abundance in OE group than in control group, were protein candidates that specifically interacted with circERBB2. c RNA pulldown followed by western blot confirmed interaction between circERBB2 and PA2G4. d RIP followed by qPCR confirmed interaction between circERBB2 and PA2G4. 18S and ERBB2 mRNA were used as negative control (n = 3). e FISH+IF double staining showed co-localization of circERBB2 (green) and PA2G4 (red) in the nucleolus. Co-localization was indicated by arrowhead. Scale bar, 10 μm. f PA2G4 was effectively silenced by 2 siRNAs. β-actin was used as internal control. g CCK8 assay showed that silencing of PA2G4 impaired proliferation of SGC-996 cells (n = 3). h qPCR assay showed that silencing of PA2G4 decreased steady level of 45S pre-rRNA (n = 3). i Silencing of PA2G4 decreased rDNA promoter activity (n = 3). j Co-IP followed by western blot confirmed interaction between PA2G4 and TIFIA. k IF double staining showed co-localization of TIFIA (green) and PA2G4 (red) in the nucleolus. Co-localization was indicated by arrowhead. Scale bar, 10 μm. l OE of circERBB2 significantly increased interaction between TIFIA and Pol I, and silencing of circERBB2 decreased interaction between TIFIA and Pol I. Quantitative data from three independent experiments was presented as mean ± SD (error bars). P-values were determined by paired, two-tailed two sample t-test. *: p < 0.05; **: p < 0.01