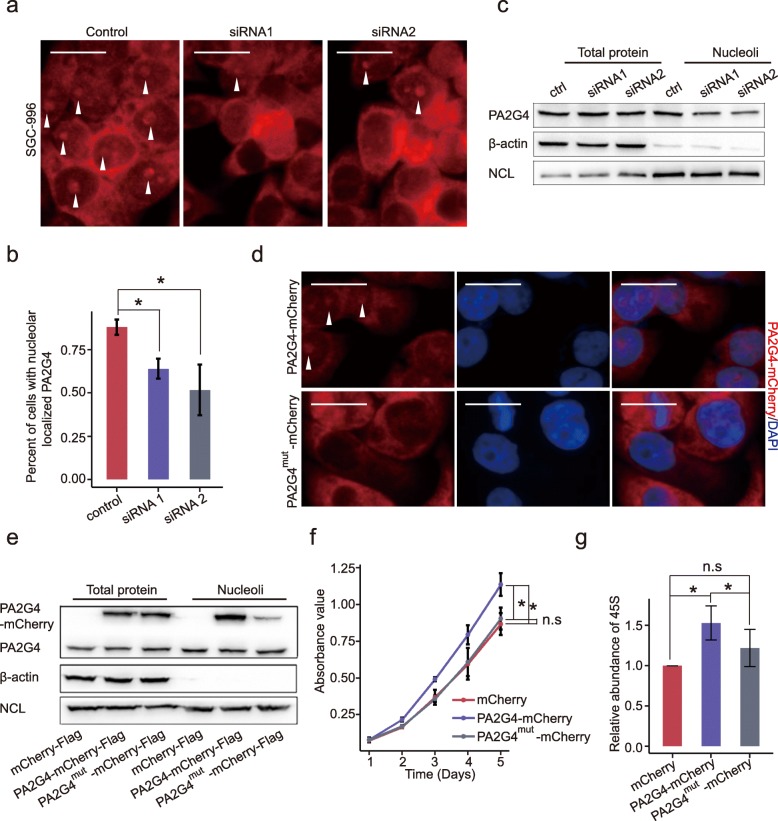
Fig. 6.
circERBB2 regulates nucleolar-localization of PA2G4. a IF assay showed that silencing of circERBB2 decreased nucleolar localization of PA2G4. Arrowhead showed nucleoli stained with PA2G4. Scale bar: 20 μm. b Quantitative analysis of the percentage of cells with nucleolar enrichment of PA2G4 (n = 4). c Western blot showed that silencing of circERBB2 had no effect on total amount PA2G4, but significantly decreased nucleolar localized PA2G4. β-actin and NCL were used as internal control for total and nucleolar protein, respectively. d IF showed that mutation of polybasic region impaired nucleolar enrichment of PA2G4. Arrowhead showed nucleoli stained with PA2G4. Scale bar: 20 μm. e Western blot showed that mutation of polybasic region impaired nucleolar enrichment of PA2G4. Distribution of endogenous PA2G4 was unaffected. f CCK8 assay showed that PA2G4-mCherry, but not PA2G4mut-mCherry, effectively promoted proliferation of SGC-996 cells (n = 3). g PA2G4-mCherry, but not PA2G4mut-mCherry, promoted rDNA transcription in GBC cells (n = 3). Quantitative data from three independent experiments was presented as mean ± SD (error bars). P-values were determined by paired, two-tailed two sample t-test. *: p < 0.05; n.s: not significant