Abstract
Variants in DONSON were recently identified as the cause of microcephaly, short stature, and limb abnormalities syndrome (MISSLA). The clinical spectra of MISSLA and Fanconi anaemia (FA) strongly overlap. For that reason, some MISSLA patients have been clinically diagnosed with FA. Here, we present the clinical data of siblings with MISSLA featuring a novel DONSON variant and summarize the current literature on MISSLA. Additionally, we perform computer-aided image analysis using the DeepGestalt technology to test how distinct the facial features of MISSLA and FA patients are. We show that MISSLA has a specific facial gestalt. Notably, we find that also FA patients feature facial characteristics recognizable by computer-aided image analysis. We conclude that computer-assisted image analysis improves diagnostic precision in both MISSLA and FA.
Subject terms: Genetics research, Translational research
Introduction
Since 30–40% of all patients with genetic disorders show characteristic craniofacial features [1], a detailed examination of the facial gestalt is important when phenotyping patients with suspected genetic disorders. However, the gestalt examination depends on the expertise of the examining physician [2]. As an additional approach, modern computer-aided image analysis programmes such as Face2Gene (FDNA Inc., Boston, USA) offer machine-learning-based algorithms combining facial recognition software with clinical knowledge to evaluate two-dimensional patient images (i.e. photographs) [3, 4]. Basel-Vanagaite et al. [3] have been the first to show that this may be an effective tool in the diagnosis of rare genetic disorders. In molecularly proven cases of Cornelia-de-Lange syndrome the approach has a diagnostic accuracy similar to clinicians [3]. Comparable results have been reported in other disorders with facial dysmorphisms [2, 5–18] suggesting the technology complements conventional phenotyping, and improves clinical diagnosis and laboratory analysis [19].
Microcephaly, short stature and limb abnormalities syndrome (MISSLA) is a rare dysmorphic autosomal recessive disorder caused by variants of the Downstream neighbour of SON (DONSON) [20–24]. As a component of the replisome, DONSON plays an important function in the stabilization of the replication fork [22, 23, 25]. DONSON promotes the activation of intra-S-phase and G2/M-phase checkpoints for the correction of replication errors. DONSON, thus, protects the nucleus against replication stress. The rate of cell proliferation is slowed by a reduced or aberrant DONSON expression [22]. This explains some of the clinical signs, such as severe intrauterine and postnatal growth retardation or severe microcephaly. MISSLA includes anomalies of the extremities, such as mesomelia of all four extremities or radial abnormalities, as well as a craniofacial dysmorphism, which is characterized by a short palpebral fissure, microstomia, micrognathia, and a broad nose with a prominent bridge. Affected individuals often die perinatally due to incomplete lung development. A neuronal function was implied by altered movement patterns of Caenorhabditis elegans mutants featuring DONSON variants [26]. Since microcephaly, pre- and postnatal growth retardation, and skeletal abnormalities, especially radial ray defects, occur in both diseases some MISSLA patients have originally been diagnosed with Fanconi anaemia (FA) [21, 24]. FA is heterogeneous with causative variants in 21 genes that are—just like DONSON—involved in the repair of DNA damage [27]. The cellular phenotype of hypersensitivity to DNA interstrand crosslinking agents is also observed in MISSLA patients suggesting functional overlap of the involved genes [24, 28]. Notably, DONSON is known to act in the ATM- and RAD3-related (ATR)-dependent pathway [22] of which the FA proteins are substrates [29].
Features of FA not yet described in MISSLA include malformations of the heart and kidneys, and progressive bone marrow failure, as well as a significantly increased risk of malignancy [28, 30–33]. Characteristic facial features of FA are microcephaly and, in some cases, small eyes/microphthalmia [30].
Here, we present the clinical manifestations of two MISSLA siblings featuring a novel likely pathogenic DONSON variant. We also test the ability of Face2Gene RESEARCH to distinguish MISSLA from FA.
Patients and methods
Image analysis
We developed classifiers for the gestalt of MISSLA syndrome and FA analogous to the DeepGestalt technology. For the development and evaluation of classifiers based solely on the image data set described below, we used the Face2Gene RESEARCH application (www.face2gene.com). The underlying methodology of DeepGestalt has been recently described at length [4].
Acquisition of MISSLA and FA cases
Pubmed and the Online Mendelian Inheritance in Man (OMIM) database were searched for images of MISSLA and FA patients suitable for this study.
Pubmed was searched with the following medical subject headings (meSH terms) (including the different names of a syndrome caused by DONSON variants):
(“Fanconi Anemia”[Mesh]) AND (“Case Reports”[Publication Type] OR “Review”[Publication Type])
(DONSON[Title] OR MISSLA[Title] OR MIMIS[Title] OR MMS[Title] OR Microcephaly Micromelia[Title] OR Microcephaly Short Stature Limb Abnormality[Title] OR Ives-Houston[Title]) AND (Case Reports[Publication Type] OR Review[Publication Type])
Nineteen suitable FA patient photos of 18 patients were found from 12 case reports and reviews (Suppl. Table 1) via Pubmed. Two images of two FA patients from Wiedemanns Atlas klinischer Syndrome [34] were added to build the final FA cohort.
The query yielded no suitable MISSLA patient images. Therefore, images were taken from the publications listed in the MISSLA-related OMIM entries yielding eight images of unrelated individuals. We added one image of the patient originally published by Schulz et al. [24] now depicted at the age of 2 years (Suppl. Fig. 1). The two images of the siblings reported here were not recognized by Face2Gene due to image quality reasons (closed eyes, non-frontal positioning of the head). The parents gave written informed consent for the publication of their children’s photos and data.
Patient 1
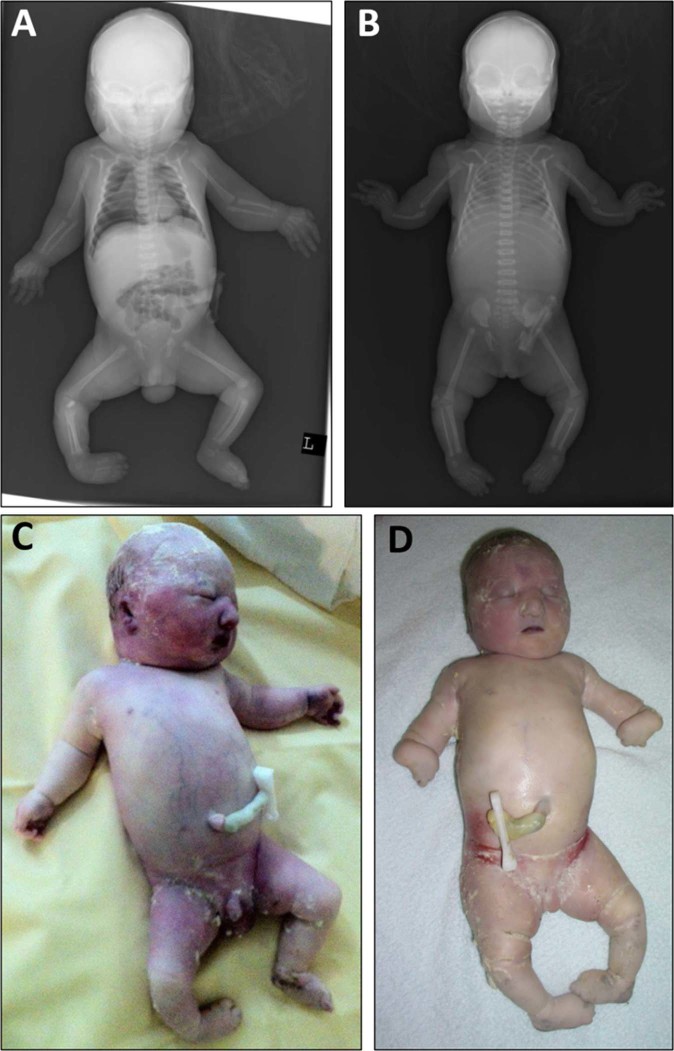
Patient 1 (Fig. 1a, c, Table 1) was a male child, born to healthy non-consanguineous parents of German descent at 38 weeks gestation. Prenatal ultrasound showed short long bones and ribs, club feet, thickened nuchal fold, and cleft palate. He had two healthy older sisters. He died directly after birth due to an untreatable lung hypoplasia. Birth length was 46 cm (−2.3 SD), weight 1960 g (−2.7 SD), OFC was 29 cm (−5.6 SD). Hypospadias was noted. Facial anomalies include broad nose with hypoplastic nostrils, microstomia, retrognathia, dysplastic ears, full cheeks, and short neck. Parents declined a post mortem examination, but a radiograph suggested a diaphragmatic hernia.
Fig. 1.
Babygrams of a patients 1 and b 2 note bilateral shortened radius and ulna, hypoplasia of thumbs and club feet in patient 1, as well as bilateral radial and thumb aplasia and club feet in patient 2. Photos of c patients 1 and d 2, note the facial features of both newborns: short palpebral fissures, a broad nose with hypoplastic nostrils, small mouth, and retrognathia, characteristic for MISSLA
Table 1.
Clinical features of published MISSLA patients
| This study | Schulz et al. (2018) | Evrony et al. (2017) | Reynolds et al. (2017) | Milner et al. (1992) | Ives and Houston (1980) | Summary | |
|---|---|---|---|---|---|---|---|
| Number of patients | 2 | 2 | 12 | 26 | 3 | 14 | 59 |
| Number of families | 1 | 1 | 9 | 8 | 1 | 1 | 21 |
| Origin | German | German | First Nations Community | Italian, English (UK), Somalian, American (USA), Turkish, Indian, Saudi Arabian | Palestinian | Cree Indians (First Nations Community) | |
| Gender | 1 F/1 M | 2 F/0 M | 8 F/4 M | 12 F/14 M | 2 F/1 M | 14 NA | 25 F/20 M, 14 NA |
| Consanguinity | 0/2 | 0/2 | 12 NA “most individuals“ | 9/9 17 NA | 3/3 | 14/14 | 26/30, 29 NA |
| Intrauterine growth restriction | 2/2 | 2/2 | 9/9, 3 NA | 12/15, 11 NA | 2/3 | 14/14 | 41/45, 14 NA |
| Short stature | 2/2 | 2/2 | 9/9, 3 NA | 20/23, 3 NA | 2/3 | 14/14 | 49/53, 6 NA |
| Microcephaly | 2/2 | 2/2 | 9/9, 3 NA | 17/18, 8 NA | 2/3 | 14/14 | 46/48, 11 NA |
| Broad nose | 2/2 | 1/2 | 1/1, 11 NA | 6/6, 20 NA | 3/3 | 14/14 | 27/28, 31 NA |
| Other facial dysmorphisms | 2/2 | 2/2 | 1/1, 11 NA “Nearly all” | 11/11, 15 NA | 3/3 | 14/14 | 33/33, 26 NA |
| Radius abnormalities | 2/2 (1 bil aplasia, 1 bil hypoplasia) | 2/2 (1 unil dysplasia and contral aplasia, 1 bil dysplasia) | 12 NA “Frequent absence” | 3/3 (1 radial head and humerus synostosis, 1 bil hypoplasia, 1 aplasia) 23 NA | 2/3 (radial head dislocation: 1 bil, 1 unil) | 6/6, 8 NA | 15/16, 43 NA |
| Thumb abnormalities | 2/2 (1 bil aplasia, 1 bil hypoplasia) | 2/2 (hypoplasia: 1 unil, 1 bil) | 1/1 aplasia, 11 NA | 1/3 bil aplasia, 23 NA | 2/3 bil dysplasia | 6/6, 8 NA | 14/17, 42 NA |
| Mesomelia | 2/2 | 2/2 | 12 NA | 2/4, 22 NA | 3 NA | 14/14 | 20/22, 37 NA |
| Other skeletal abnormalities | 2/2 | 2/2 | 12 NA | 17/19, 7 NA | 3/3 | 6/6, 8 NA | 29/32, 27 NA |
| Perinatal death | 2/2 | 0/1, 1 NA | 12 NA | 2/19, 7 NA | 0/3 | 14/14 | 18/39 20 NA |
| Other | Hypoplastic lung | Foetal MRI: flat sulcus centralis and corpus callosum agenesis lissencephaly | 2 simplified gyral pattern, hypoplastic or absent corpus callosum, | PFO | Hypopigmented maculae | 3 corpus callosum agenesis |
NA not applicable; SNV single nucleotide variant; F female; M male; unil, unilateral; bil, bilateral; contral, contralateral; PFO patent foramen ovale
Patient 2
The younger sister of patient 1 (Fig. 1b, d; Table 1) featured severe intrauterine growth retardation, with shortened thin long bones and ribs and club feet. A thickened nuchal fold and a cleft palate were present. She was born at 38 weeks gestation. Birth length was 40 cm (−4.9 SD), weight 1590 g (−3.6 SD), OFC was 26.5 cm (−10.6 SD). She died due to untreatable lung hypoplasia, too. Notably, observations were bilateral aplasia of the radius and thumb with bowed ulnas, cutaneous syndactyly of the fingers II–V and toes II–IV and a sandal gap. The following facial features were notable: short and downslanted palpebral fissure, micrognathia, microstomia, broad nose with hypoplastic nostrils, short neck, and dysplastic ears. Investigation by ultrasound, however, showed atresia of the right choana and both external auditory canals. Parents again declined a post mortem examination.
Analysis by a 1M oligo aCGH (Agilent Technologies) was unremarkable. The siblings’ phenotype prompted Sanger sequencing of the protein coding part and flanking bases of DONSON. This revealed compound heterozygosity for the two variants c.[661T>C];[1433C>T] in exons 4 and 9, respectively (Suppl. Fig. 2) in patient 2. The parents were confirmed to be heterozygotes. No material from patient 1 was available for testing (all variants described in this paper refer to the DONSON transcript NM_017613.3 positioned at NC_000021.8:g.34949859-34961267, exons and introns are numbered as with Reynolds et al. (2017) and Evrony et. al (2017)).
The variant c.661T>C (p.Trp221Arg) has not previously been reported in MISSLA. However, the variant in trans c.1433C>T (p.Pro478Leu) has previously been described in MISSLA [24] (American College of Medical Genetics (ACMG) criterion PM3). c.661T>C (p.Trp221Arg) is neither found in the Exome Aggregation Consortium’s (ExAC) database [35] nor in the database of the 1000 Genomes Project [36] (ACMG criterion PM2). Trp221 is highly conserved (Suppl. Fig. 2B), with a phyloP [37] score of 5.1 and this variant was predicted to be disease causing by MutationTaster [38] (ACMG criterion PP3). Given the patients’ highly MISSLA suggestive phenotype (e.g. skeletal malformations, lethal lung hypoplasia) and family history (ACMG criterion PP4) we classified c.661T>C (p.Trp221Arg) as “likely pathogenic” according to the ACMG guidelines for the interpretation of sequence variants [39] (2PM + 2PP). Variant and phenotypic data were submitted to the Leiden Open Variation Database (www.LOVD.nl/DONSON) Individual ID # 00207473.
Control cohorts
To test the ability of Face2Gene RESEARCH to recognize MISSLA and FA we constructed three control image cohorts (Smith-Lemli-Opitz syndrome (SLOS), dysmorphic and non-dysmorphic).
Dysmorphic and Smith-Lemli-Opitz control cohorts
We searched the archive of the Institute of Medical Genetics and Human Genetics of the Charité Berlin for photos of patients with a molecularly confirmed diagnosis who did not have FA, MISSLA or SLOS and consented to the use of their images for scientific purposes. A maximum of two cases of the same family was used. One hundred and eighty-three images of 116 patients featuring 88 different syndromes were, eventually, used for the cohort of dysmorphic patients (Suppl. Table 1). Eighty images of 65 SLOS patients from of the SLOS cohort originally used by Pantel et al. [13] were used as an additional control cohort (Suppl. Table 1) [13].
Non-dysmorphic control cohort
To build a cohort of non-dysmorphic individuals, we searched online for 116 publicly accessible portrait photos matching the 116 cases of the dysmorphic control cohort regarding age, gender and ethnicity (Suppl. Table 1). Matching was performed because other factors than facial dysmorphism (such as ethnicity) may confound Face2Gene [2, 13]. Absence of facial dysmorphism was evaluated by at least two clinicians.
Suitable portrait photos
Portrait photos had to meet the following criteria: a picture had to feature an individual confirmed to have one of the disorders analysed in this study, the patient should not have any other genetic disorder and the entire face of the patient (hairline, both eyes, nose, mouth and chin) had to be recognizable (see Suppl. Fig. 1 for an example). If necessary, photos were manually trimmed prior to analysis by Face2Gene.
Image classification and statistical analysis
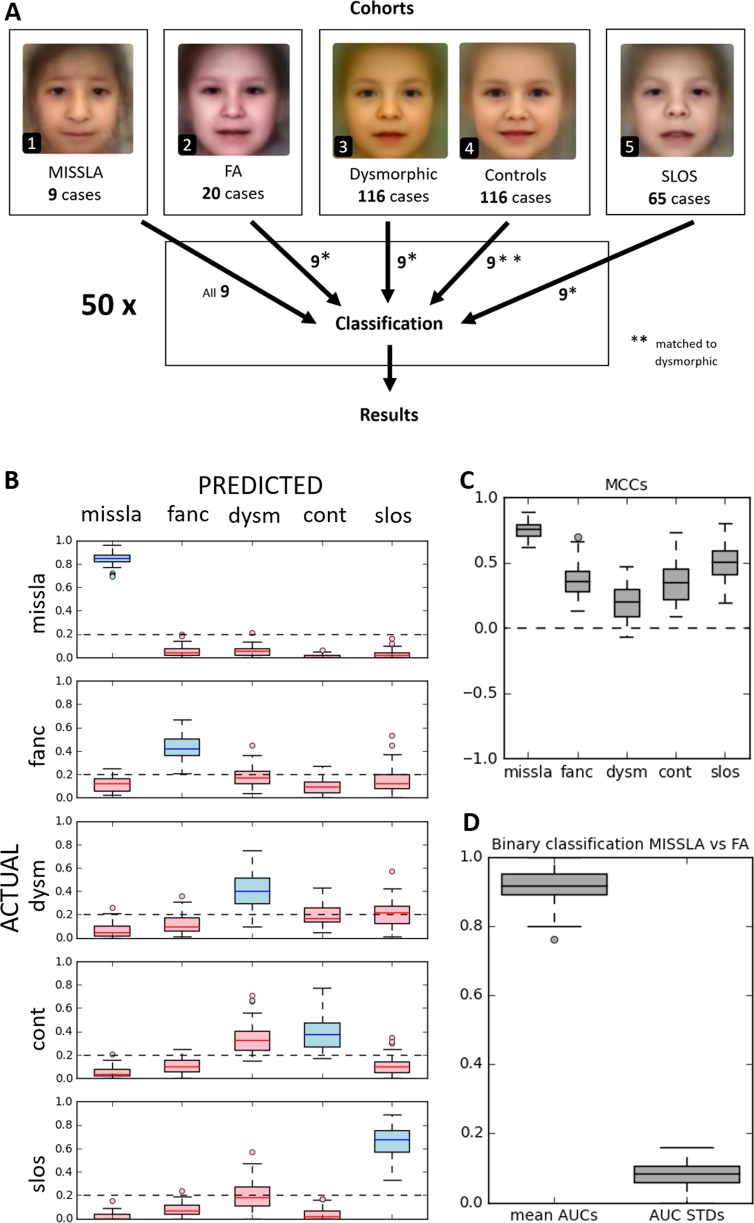
Image classifications were performed as described by Liehr et al. [8] with the Face2Gene RESEARCH application (v. 19.1.0). Cross-validation was performed according to the default settings of the application (method: holdout, splits: 10). Next to Face2Gene CLINIC which evaluates patient photos using the pretrained neural network DeepGestalt [4], Face2Gene RESEARCH enables the design of user-specific classifiers. When the Face2Gene RESEARCH application is trained with different numbers of cases per class, this may bias the classifier [13]. As the MISSLA cohort was the smallest consisting of just nine images, we used these nine images and randomly selected nine cases of each of the other cohorts to run a classification experiment. This process was repeated 50 times (Fig. 2a), in order to minimize a potential bias of the nine images specifically selected per cohort.
Fig. 2.
a Study design, composite syndrome images of (A1) MISSLA, (A2) FA, (A3) dysmorphic control, (A4) non-dysmorphic control, and (A5) SLOS control individuals. b Boxplot of the confusion matrix results. Rows depict actual classes; columns depict predicted classes. Correct classifications are shown in blue, false classifications in red. Dashed line indicates 20% of accuracy (expected for random chance). Note: all mean true positive rates above 20%. c Matthew’s Correlation Coefficients (MCCs). Dashed line indicates 0 (expected for random chance). Note: all mean MCCs above 0. d Results of the binary classification. Distribution of the AUCs under the receiver operating characteristic curves and their standard deviations
Face2Gene’s accuracy of the binary comparison of MISSLA and FA cohorts was assessed by measuring the mean area under the curve (AUC) of the receiver operating characteristic curve. The power to compare and separate all five classes (MISSLA, FA, SLOS, dysmorphic, non-dysmorphic controls) was evaluated by measuring true positive rates (TPRs), false positive rates (FPRs) and calculating Matthews Correlation Coefficient (MCC) defined as
with TP = true positives, TN = true negatives, FP = false positives, FN = false negatives.
A value of 1 reflects a perfect classification, a value of 0 a purely random result and a negative value a result worse than that of a random classifier.
To test a possible overfitting of the RESEARCH application, we also performed 50 classification experiments randomly labeling the 9 cases of each class of the actual experiments to one of 5 classes (Suppl. Fig. 3). Results were compared to the 50 actual experiments using a two-sided t-test.
Results
Gestalt composite images
To visualize the average appearance of the individuals of our cohorts, we created composite images using Face2Gene RESEARCH (Fig. 2a1–a5). The MISSLA composite shows distinct facial features, such as a broad nose with underdeveloped nasal alae, short and downslanted palpebral fissures, and microstomia. The FA composite indicates small eyes and microstomia. The dysmorphic composite shows no specific facial appearance, as it contains a wide number of different genetic disorders. The SLOS composite depicts anteverted nares a broad nasal bridge and ptosis.
Multiclass comparison
To determine the distinguishability of MISSLA syndrome and FA we performed a computer-aided image classification of MISSLA, FA, SLOS and dysmorphic patients, and non-dysmorphic control individuals using the Face2Gene RESEARCH software. Notably, all median TPRs were above 20% (p < 0.05), i.e. better than the expected value for an evenly distributed result achieved by a 5-class random classifier (Fig. 2b). The highest TPRs were seen for the MISSLA cohort with a median TPR of 84% (p = 2.7 ×10−49). The median FPRs of other cohorts wrongly classified as MISSLA are lower than 20% (Fig. 2b) (p < 0.05). Remarkably, on average, only 6% of the MISSLA images were classified as FA (p = 9.9 ×10−13).
The second highest median TPR (66%, p = 1.3 × 10−32) was observed in the SLOS cohort, Notably, the FA cohort’s TPR amounted to 44% (p = 6.4 × 10−20), indicating distinct facial features in some of the patients of the FA cohort.
Interestingly, of the 20 median FPRs only the rate of non-dysmorphic images classified as dysmorphic was significantly higher than expected by random chance (34%, p = 1.7 ×10−7) (Fig. 2b).
All five MCCs show positive values (Fig. 2c), indicating a classification better than random chance (p < 0.05). The highest MCCs were found for the class MISSLA (median MCC: 0.75, p = 3.0 × 10−52) the lowest MCCs for the dysmorphic class (median MCC: 0.19, p = 42.7 × 10−12).
Binary comparison
To directly test the distinguishability of MISSLA and FA, we also used the RESEARCH app for a binary comparison of the two cohorts. Mean AUCs were particularly high (mean 0.91, p = 1.6 ×10−21) with only low standard deviations (Fig. 2d), suggesting that the two syndromes differ in their facial appearance.
Discussion and conclusion
Here, we report the clinical and molecular data of two so far unpublished siblings with MISSLA, caused by the known disease-causing variant c.1433C>T (p.Pro478Leu) and the novel variant c.661T>C;(p.Trp221Arg) in DONSON. We summarize the phenotypes of all previously published MISSLA cases and present the first study to use a computer-aided image analysis approach to distinguish MISSLA and FA.
MISSLA and FA can be differentiated by computer-aided image analysis
Despite phenotypic overlap, our results reveal that MISSLA has a typical face that can be clearly distinguished from FA by computer-assisted image analysis. The high TPRs and MCCs for MISSLA and FA also indicate that they differ from the other control groups. Whether FA has specific facial characteristics is sometimes debated. Our work supports the assumption of Avila et al. [40] that there is a characteristic facial gestalt of FA. It is, however, possible that the TPRs of FA are lower than those of MISSLA and SLOS because not all FA patients feature these characteristic facial features. Knaus et al. [11] showed that DeepGestalt can classify patients of the same phenotypic series according to their mutated gene. Our FA cohort is based on FA patients featuring variants in different genes, thus specific facial features may only be caused by some of these.
Notably, in their investigation of Emmanuel and Pallister-Killian syndromes, Liehr et al. [8] reported better differentiation between dysmorphic and non-dysmorphic cases than we have reported here. This might be due to the following reasons: (a) they used more images than we did, (b) their data set consisted of four not five classes, (c) their cohorts differed in the numbers of images used to build them, which according to Pantel et al. [13] may confound the classification process, and (d) their cohorts were not matched regarding age, gender, and ethnicity which could have confounded classification, too [2].
Pantel et al. [13] have demonstrated that more than nine images are required to build the best possible DeepGestalt-classifier for a given syndrome. Unfortunately, the number of published images of unrelated MISSLA patients, suitable for Face2Gene, is just 9. This shows the need for scientifically monitoring patients with rare, clinically variable diseases like MISSLA, and to share the findings.
MISSLA is not a new subtype of FA
One reason to assume that MISSLA represents a specific disease entity were mitomycin C test results. In contrast to FA, Milner et al. [21] found no increased number of chromosomal aberrations after mitomycin C testing of MISSLA patients. However, Evrony et al. [23] and Schulz et al. [24] did find those. Some patients were even originally diagnosed with FA. This raised the question whether regarding the strong clinical overlap and similarities in the cellular phenotype MISSLA should be seen as a new subtype of FA.
Since haematological symptoms, commonly seen in FA patients, have not been described in MISSLA, since heterozygote DONSON variant carriers have not been associated with an increased tumour risk, since there are specific differences in skeletal malformations between MISSLA and FA, and in particular since MISSLA has a characteristic facies, we still assume that it is a specific clinical entity.
Further research necessary
Face2Gene RESEARCH is supposed to work with images of different quality. We, however, did not test this.
Further research is also needed to determine the potential of other image analysis programmes, or a combination of such programmes for the distinction of MISSLA and FA. Although achieving a certain precision in the identification of MISSLA, FA, and other syndromes, our results are improvable. Our main obstacle was the limited number of suitable photos. The control experiments’ results make an overfitting of Face2gene RESEARCH to our limited data set unlikely. However, a larger number of photos would enable more detailed investigation of the facial MISSLA phenotype and analysis of a potential facial genotype–phenotype correlation in FA patients. The classifiers presented in this study did not reach the accuracy of a clinician in differentiating dysmorphic and non-dysmorphic faces.
However, we conclude computer-assisted image analysis can support the subjective task of classical clinical diagnosis, potentially helping to identify MISSLA among clinically diagnosed FA patients who were not genetically confirmed.
Supplementary information
Acknowledgements
We thank the families for their cooperation and Carola Dietrich, Nicole Fleischer and Peter Krawitz for technical support. MAM is participant in the BIH Charité Junior Clinician Scientist Program funded by the Charité—Universitätsmedizin Berlin and the Berlin Institute of Health.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version of this article (10.1038/s41431-019-0469-3) contains supplementary material, which is available to authorized users.
References
- 1.Hart TC, Hart PS. Genetic studies of craniofacial anomalies: clinical implications and applications. Orthod Craniofac Res. 2009;12:212–20. doi: 10.1111/j.1601-6343.2009.01455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lumaka A, Cosemans N, Lulebo Mampasi A, Mubungu G, Mvuama N, Lubala T, et al. Facial dysmorphism is influenced by ethnic background of the patient and of the evaluator. Clin Genet. 2017;92:166–71. doi: 10.1111/cge.12948. [DOI] [PubMed] [Google Scholar]
- 3.Basel-Vanagaite L, Wolf L, Orin M, Larizza L, Gervasini C, Krantz ID, et al. Recognition of the Cornelia de Lange syndrome phenotype with facial dysmorphology novel analysis. Clin Genet. 2016;89:557–63. doi: 10.1111/cge.12716. [DOI] [PubMed] [Google Scholar]
- 4.Gurovich Y, Hanani Y, Bar O, Fleischer N, Gelbman D, Basel-Salmon L, et al. Identifying facial phenotypes of genetic disorders using deep learning. Nat Med. 2019;25:60–64. doi: 10.1038/s41591-018-0279-0. [DOI] [PubMed] [Google Scholar]
- 5.Gripp KW, Baker L, Telegrafi A, Monaghan KG. The role of objective facial analysis using FDNA in making diagnoses following whole exome analysis. Report of two patients with mutations in the BAF complex genes. Am J Med Genet A. 2016;170:1754–62. doi: 10.1002/ajmg.a.37672. [DOI] [PubMed] [Google Scholar]
- 6.Gardner OK, Haynes K, Schweitzer D, Johns A, Magee WP, Urata MM, et al. Familial recurrence of 3MC syndrome in consanguineous families: a clinical and molecular diagnostic approach with review of the literature. Cleft Palate Craniofac J. 2017;54:739–48. doi: 10.1597/15-151. [DOI] [PubMed] [Google Scholar]
- 7.Graul-Neumann LM, Mensah MA, Klopocki E, Uebe S, Ekici AB, Thiel CT, et al. Biallelic intragenic deletion in MASP1 in an adult female with 3MC syndrome. Eur J Med Genet. 2018;61:363–8. doi: 10.1016/j.ejmg.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 8.Liehr T, Acquarola N, Pyle K, St-Pierre S, Rinholm M, Bar O, et al. Next generation phenotyping in Emanuel and Pallister-Killian syndrome using computer-aided facial dysmorphology analysis of 2D photos. Clin Genet. 2018;93:378–81. doi: 10.1111/cge.13087. [DOI] [PubMed] [Google Scholar]
- 9.Hadj-Rabia S, Schneider H, Navarro E, Klein O, Kirby N, Huttner K, et al. Automatic recognition of the XLHED phenotype from facial images. Am J Med Genet A. 2017;173:2408–14. doi: 10.1002/ajmg.a.38343. [DOI] [PubMed] [Google Scholar]
- 10.Valentine Matthew, Bihm Dustin C.J., Wolf Lior, Hoyme H. Eugene, May Philip A., Buckley David, Kalberg Wendy, Abdul-Rahman Omar A. Computer-Aided Recognition of Facial Attributes for Fetal Alcohol Spectrum Disorders. Pediatrics. 2017;140(6):e20162028. doi: 10.1542/peds.2016-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knaus A, Pantel JT, Pendziwiat M, Hajjir N, Zhao M, Hsieh T-C, et al. Characterization of glycosylphosphatidylinositol biosynthesis defects by clinical features, flow cytometry, and automated image analysis. Genome Med. 2018;10:3. doi: 10.1186/s13073-017-0510-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zarate YA, Smith-Hicks CL, Greene C, Abbott M-A, Siu VM, Calhoun ARUL, et al. Natural history and genotype-phenotype correlations in 72 individuals with SATB2-associated syndrome. Am J Med Genet A. 2018;176:925–35. doi: 10.1002/ajmg.a.38630. [DOI] [PubMed] [Google Scholar]
- 13.Pantel JT, Zhao M, Mensah MA, Hajjir N, Hsieh T-C, Hanani Y, et al. Advances in computer-assisted syndrome recognition by the example of inborn errors of metabolism. J Inherit Metab Dis. 2018;41:533–9. doi: 10.1007/s10545-018-0174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira CR, Altassan R, Marques-Da-Silva D, Francisco R, Jaeken J, Morava E. Recognizable phenotypes in CDG. J Inherit Metab Dis. 2018;41:541–53. doi: 10.1007/s10545-018-0156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Y, Wangler MF, McGuire AL, Lupski JR, Posey JE, Khayat MM, et al. The phenotypic spectrum of Xia-Gibbs syndrome. Am J Med Genet A. 2018;176:1315–26. doi: 10.1002/ajmg.a.38699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vorravanpreecha N, Lertboonnum T, Rodjanadit R, Sriplienchan P, Rojnueangnit K. Studying Down syndrome recognition probabilities in Thai children with de-identified computer-aided facial analysis. Am J Med Genet A. 2018;176:1935–40. doi: 10.1002/ajmg.a.40483. [DOI] [PubMed] [Google Scholar]
- 17.Mishima Hiroyuki, Suzuki Hisato, Doi Michiko, Miyazaki Mutsuko, Watanabe Satoshi, Matsumoto Tadashi, Morifuji Kanako, Moriuchi Hiroyuki, Yoshiura Koh-ichiro, Kondoh Tatsuro, Kosaki Kenjiro. Evaluation of Face2Gene using facial images of patients with congenital dysmorphic syndromes recruited in Japan. Journal of Human Genetics. 2019;64(8):789–794. doi: 10.1038/s10038-019-0619-z. [DOI] [PubMed] [Google Scholar]
- 18.Carli D, Giorgio E, Pantaleoni F, Bruselles A, Barresi S, Riberi E, et al. NBAS pathogenic variants: defining the associated clinical and facial phenotype and genotype-phenotype correlations. Hum Mutat. 2019;40:721–8. doi: 10.1002/humu.23734. [DOI] [PubMed] [Google Scholar]
- 19.Hsieh T-C, Mensah MA, Pantel JT, Aguilar D, Bar O, Bayat A, et al. PEDIA: prioritization of exome data by image analysis. Genet Med. 2019. 10.1038/s41436-019-0566-2 [DOI] [PMC free article] [PubMed]
- 20.Ives EJ, Houston CS. Autosomal recessive microcephaly and micromelia in Cree Indians. Am J Med Genet. 1980;7:351–60. doi: 10.1002/ajmg.1320070317. [DOI] [PubMed] [Google Scholar]
- 21.Milner RD, Khallouf KA, Gibson R, Hajianpour A, Mathew CG. A new autosomal recessive anomaly mimicking Fanconi’s anaemia phenotype. Arch Dis Child. 1993;68:101–3. doi: 10.1136/adc.68.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reynolds JJ, Bicknell LS, Carroll P, Higgs MR, Shaheen R, Murray JE, et al. Mutations in DONSON disrupt replication fork stability and cause microcephalic dwarfism. Nat Genet. 2017;49:537–49. doi: 10.1038/ng.3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evrony GD, Cordero DR, Shen J, Partlow JN, Yu TW, Rodin RE, et al. Integrated genome and transcriptome sequencing identifies a noncoding mutation in the genome replication factor DONSON as the cause of microcephaly-micromelia syndrome. Genome Res. 2017;27:1323–35. doi: 10.1101/gr.219899.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schulz S, Mensah MA, de Vries H, Fröber R, Romeike B, Schneider U, et al. Microcephaly, short stature, and limb abnormality disorder due to novel autosomal biallelic DONSON mutations in two German siblings. Eur J Hum Genet. 2018;26:1282–7. doi: 10.1038/s41431-018-0128-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesly S, Bandura JL, Calvi BR, Rapid DNA. Synthesis during early embryogenesis is sensitive to maternal Humpty Dumpty protein function. Genetics. 2017;207:935–47. doi: 10.1534/genetics.117.300318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nordquist S, Smith SR, Pierce J. Systematic functional characterization of human 21st chromosome orthologs in Caenorhabditis elegans. 2017. 10.1101/136911 [DOI] [PMC free article] [PubMed]
- 27.Mamrak NE, Shimamura A, Howlett NG. Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 2017;31:93–99. doi: 10.1016/j.blre.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’Andrea AD. The Fanconi road to cancer. Genes Dev. 2003;17:1933–6. doi: 10.1101/gad.1128303. [DOI] [PubMed] [Google Scholar]
- 29.Yazinski SA, Zou L. Functions, regulation, and therapeutic implications of the ATR checkpoint pathway. Annu Rev Genet. 2016;50:155–73. doi: 10.1146/annurev-genet-121415-121658. [DOI] [PubMed] [Google Scholar]
- 30.Mehta PA, Tolar J. Fanconi anemia. In: Adam MP, Ardinger HH, Pagon RA, Wallace SE, Bean LJH, Stephens K, et al., editors. GeneReviews. Seattle, WA: University of Washington, Seattle; 2002.
- 31.Alter BP. Fanconi anemia and the development of leukemia. Best Pract Res Clin Haematol. 2014;27:214–21. doi: 10.1016/j.beha.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alter BP. Inherited bone marrow failure syndromes: considerations pre- and posttransplant. Hematol Am Soc Hematol Educ Program. 2017;2017:88–95. doi: 10.1182/asheducation-2017.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joenje H, Patel KJ. The emerging genetic and molecular basis of Fanconi anaemia. Nat Rev Genet. 2001;2:446–57. doi: 10.1038/35076590. [DOI] [PubMed] [Google Scholar]
- 34.Wiedemann H-R. Wiedemanns Atlas klinischer syndrome: Phänomenologie, Ätiologie, Differenzialdiagnose; mit 10 Tabellen. Schattauer Verlag; 2010:125.
- 35.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, 1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pollard KS, Hubisz MJ, Rosenbloom KR, Siepel A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 2010;20:110–21. doi: 10.1101/gr.097857.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 39.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Avila LF, de C, Denis Martins W, Cândido L, Ignácio SA, Bonfim CMS, et al. A study of facial pattern in patients with fanconi anemia. Cleft Palate Craniofac J. 2014;51:83–89. doi: 10.1597/11-321.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.