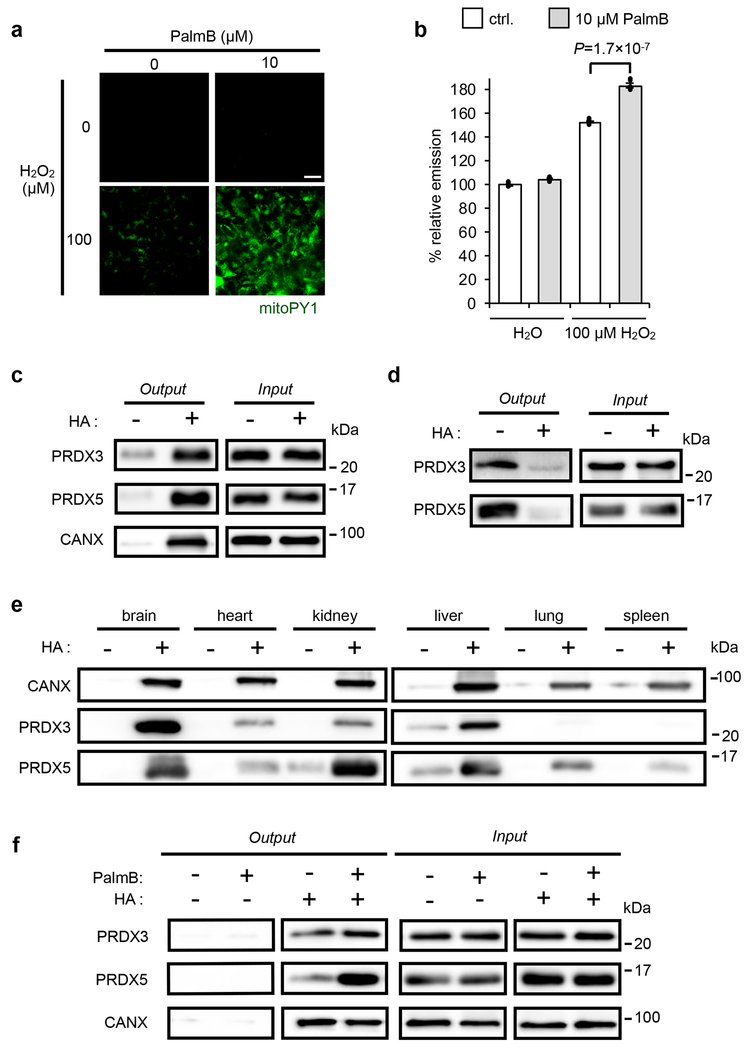
Fig. 1. Connections between S-depalmitoylation and mitochondrial redox buffering capacity.
(a) Representative images showing that PalmB treatment diminishes the mitochondrial redox buffering capacity as measured by mitoPY1 (mitochondrially targeted dye that generates H2O2-dependent fluorescence) in HEK293T cells when exposed to H2O2. 25 μm scale bar shown. (b) Quantification of the relative fluorescence intensity from mitoPY1 in each set of conditions shown in a. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n = 6 images). Data expressed as mean ± s.e.m. and normalized to control cells treated with DMSO. Dots represent individual data points. (c) ABE assay confirms S-acylation of PRDX3 and PRDX5 in HEK293T cells, with calnexin (CANX) as a known S-palmitoylated protein control. Input: total protein before enrichment; Output: S-acylated protein enriched via biotin labeled thiols upon hydroxyl amine (HA) treatment which selectively cleaves thioester bonds to expose acylated thiols. (d) Metabolic labeling with the palmitic acid analogue, 17-ODYA (50 μM) further validates PRDX3 and PRDX5 are S-palmitoylated in HEK293T cells. 17-ODYA is metabolically incorporated into palmitoylated proteins and is clicked with biotin-azide for enrichment. The signal difference between −HA and +HA lanes indicates the palmitoylation modification occurs on cysteine residues. (e) PRDX3 and PRDX5 are prone to S-acylation in vivo as determined by an ABE assay performed on mouse tissues. (f) Pan-APT inhibitor PalmB treatment increases the S-palmitoylation level of PRDX3 and PRDX5 as determined by an ABE assay in HEK293T cells. Two biological replicates performed for b-d and f.