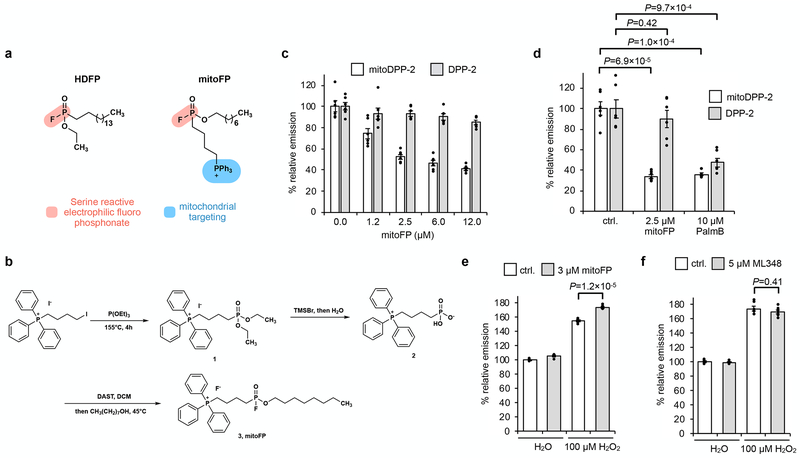
Fig. 2. Selective inhibition of mitochondrial APTs reduces mitochondrial redox buffering capacity.
(a) Design of mitoFP (right), based on the known pan-lipase inhibitor, HDFP (left). The electrophilic fluorophosphonate (red) reacts with active site serine in serine hydrolases, while the triphenylphosphonium group (blue) selectively delivers mitoFP to mitochondria. (b) Schematic for chemical synthesis of mitoFP. (c) MitoFP dose-dependent response in HEK293T cells of mitochondrial and cytosolic APT activity as measured by epifluorescence microscopy using the mitochondrial-localized APT probe, mitoDPP-2 (black), and the cytosolic APT probe, DPP-2 (grey). Data expressed as mean ± s.e.m. (n = 6 images) and normalized to control cells treated with DMSO alone. (d) Quantification of the relative fluorescence intensity from mitoDPP-2 (black) and DPP-2 (grey) in HEK293T cells comparing deactivation of mitochondrial and cytosolic APT activity, respectively, by mitoFP and PalmB. (e) Quantification of the relative fluorescence intensity from mitoPY1 showing that mitoFP diminishes the mitochondrial redox buffering capacity in HEK293T cells exposed to H2O2. (f) Quantification of the relative fluorescence intensity from mitoPY1 showing that ML348 (5 μM) does not alter mitochondrial redox buffering capacity in HEK293T cells exposed to H2O2. Statistical analyses performed with a two-tailed Student’s t-test with unequal variance (n = 6 images for d and 5 images for each e and f). Data expressed as mean ± s.e.m. and normalized to control cells treated with DMSO. Dots represent individual data points. Two biological replicates were performed for c-f.