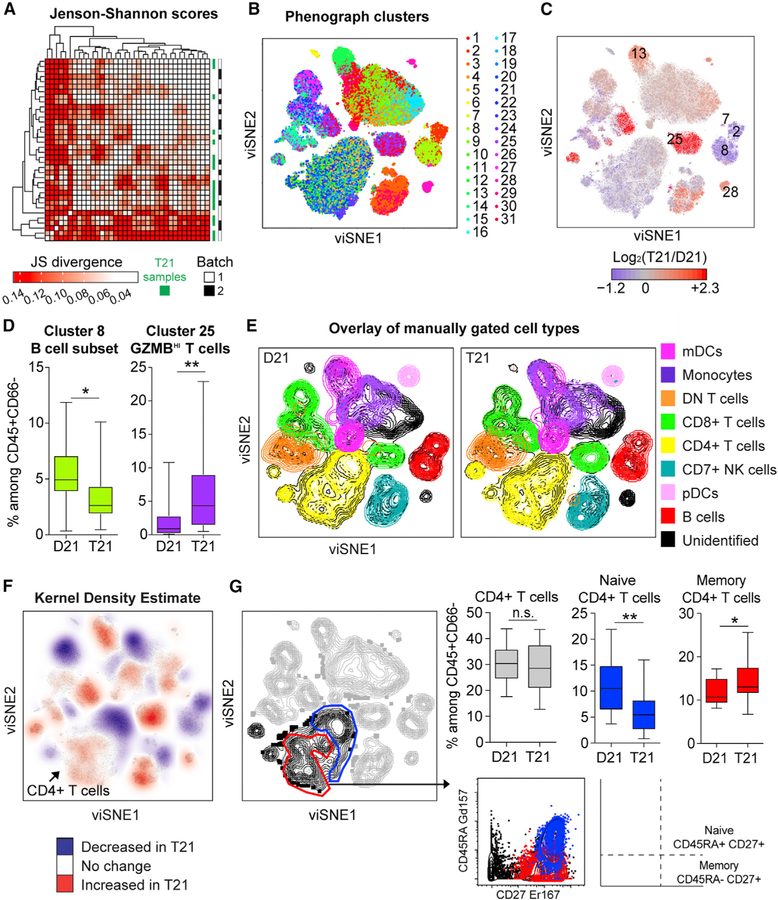
Figure 1. Mass Cytometry Reveals Global Immune Dysregulation in Adults with T21.
Intact single events enriched for non-granulocytes of hematopoietic origin (CD45+CD66−) were run through viSNE and then compared between individuals with T21 (n = 18) and D21 controls (n = 18).
(A) Similarities between overall shape of individual viSNE plots were quantified through pairwise comparisons by Jenson-Shannon (JS) scores, followed by hierarchical clustering.
(B) PhenoGraph was used to resolve 31 clusters within viSNE plots.
(C) PhenoGraph clusters were uniformly colored according to increased (red) or decreased (blue) frequency among total events in T21 viSNE plots compared to those of D21 controls. Significantly different clusters are labeled with numbers.
(D) Box and whisker plots displaying the data for PhenoGraph Clusters 8 (a subset of B cells) and 25 (a subset of T cells expressing high levels of granzyme B).
(E) Overlay of immune cell subsets defined by surface marker expression on events represented in viSNE plots.
(F) A topographical analysis, kernel density estimate (KDE), was applied to viSNE plots to visualize areas with more (red) or less (blue) event density in samples with T21. Color is overlaid on a viSNE plot showing concatenated events from all T21 samples.
(G) Manual gating of areas within viSNE plots highlighted by KDE. An area of CD4+ T cells (black) depleted of CD45RA+CD27+ events (blue) were interpreted as a subset of naive CD4+ T cells whereas an area enriched for CD45RA—CD27+ events (red) were interpreted as a subset of CD27+ memory CD4+ T cells.
In all cases, statistical significance was determined by a Student’s t test (*p ≤ 0.05 and **p ≤ 0.01). See also Figure S1. All box and whisker plots denote the median within a box extending from the 25th to 75th percentiles and error bars span minimum to maximum values within the datasets.