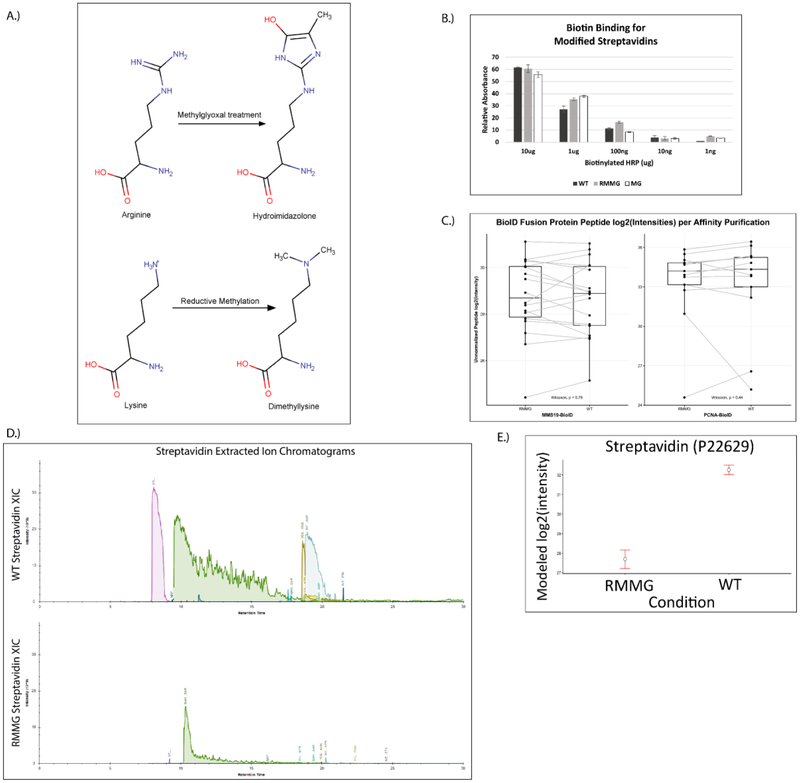
Figure 2:
Streptavidin derivatization provides protection from enzymatic digestion without disrupting biotin binding. (A) Schematic showing the derivatization of arginine and lysine residues of polypeptides to protect against enzymatic digestion. (B) Biotinylated HRP activity was measured after binding to various forms of derivatized streptavidin. HRP enzymatic activity was not impacted by the lysine and arginine derivatization, suggesting the binding of biotinylated proteins would not be disrupted. Values are given in arbitrary units of absorbance, with error bars showing the standard deviation of replicates for each bead type and HRP mass. (C) Auto-biotinylated BioID fusion proteins’ peptides are detected with similar unnormalized label-free intensities when purified by either derivatized (RMMG) or underivatized (WT) streptavidin beads within representative acquisitions. Differences between the peptide intensity populations for the BioID-MMS19 fusion protein and PCNA-BioID fusion protein are not significant between their respective RMMG and WT pulldowns by the Wilcoxon signed rank test. Box and whiskers illustrate the first and third quartiles, and the maximum and minimum observed values up to 1.5 times the interquartile range beyond the edges of the box. (D) Manually integrated extracted ion chromatograms were generated for uniquely mapping tryptic peptides derived from Streptavidin within the Skyline software. A representative pair of WT/RMMG pulldowns display the impact of derivatization on the detectable streptavidin peptide signals, dramatically reducing the overall streptavidin intensity. (E) Label-free protein intensities comparing the overall streptavidin intensity of the aggregate of all BioID data acquired here with WT beads and RMMG beads. Values displayed are modeled protein intensities, with error bars representing the reported 95% confidence intervals.