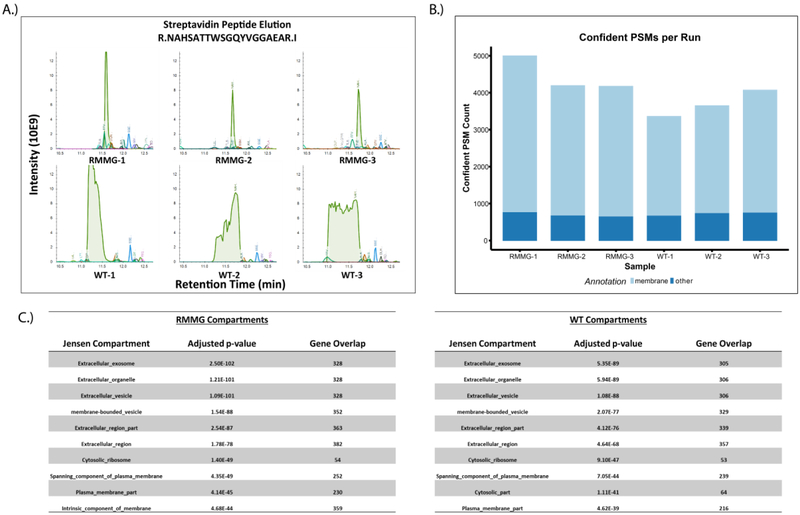
Figure 4:
Analysis of cell surface proteome using protected streptavidin beads improves peptide identification rates. (A) Extracted ion chromatogram overlay around the retention time of the streptavidin peptide NAHSATTWSGQYVGGAEAR for the cell surface biotinylation samples. As before, we observe that elution of excessive streptavidin peptides in WT runs perturbs the retention times of otherwise coeluting peptides in the RMMG runs. (B) Confident peptide spectrum matches (percolator q-value<=0.01) by sample. PSMs which mapped to protein sequences belonging to genes annotated with the membrane gene ontology cellular compartment term (GO:0016020) are shown in light blue. (C) Compartment localization enrichments were calculated from the COMPARTMENTS database through the Enrichr platform. Both RMMG and WT beads capture significant enrichment of extracellular and membrane related localizations.