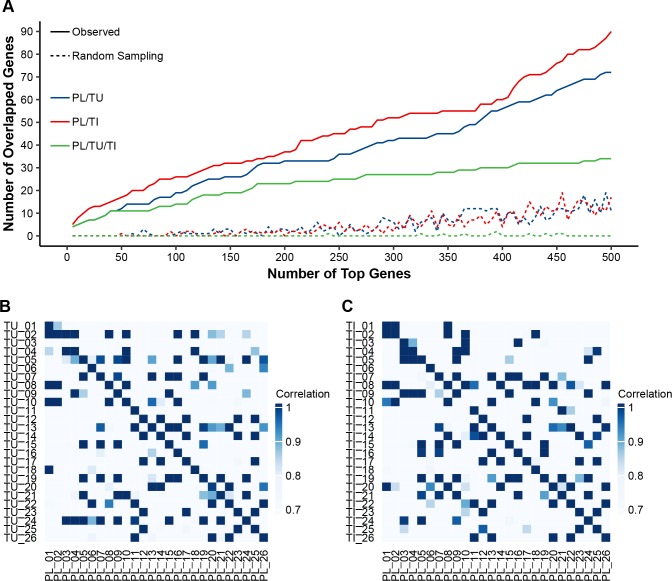
Figure 3.
Tissue relevance of the 5hmC-Seal data in HCC patient-derived cfDNA. (A) For the most variable genes in cfDNA samples, the number of overlapped genes between cfDNA and tumours (TU: blue line) or adjacent tissues (TI: red line) is significantly higher than that from random sampling (eg, hypergeometric test p<0.0001 for the top 500 modified genes in cfDNA). The green line indicates the shared genes across plasma cfDNA, tumours and adjacent tissues. (B, C) Within-subject correlation is significantly higher (Wilcoxon rank-sum test p<0.0001) between plasma cfDNA and TU/TI genomic DNA (diagonal line, mean of Pearson’s r: 0.88) than that between different individuals (mean of Pearson’s r: 0.73), based on the top 30 most variable genes in cfDNA samples in terms of 5hmC modification. cfDNA, cell-free DNA; HCC, hepatocellular carcinoma; 5hmC, 5-hydroxymethylcytosines; PL, plasma cfDNA; TI, adjacent tissue; TU, tumour.