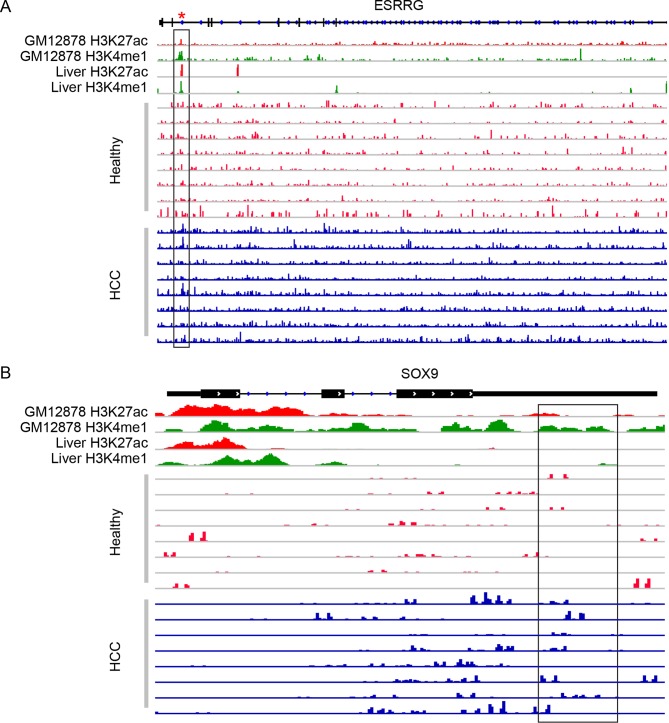
Figure 6.
Read distribution in candidate marker genes co-localised with cis-regulatory elements. The histone modification marks (H3K4me1, H3K27ac) from the ENCODE Project (GM12878) or liver tissue-derived data from the Roadmap Epigenomics Project are shown together with the 5hmC-Seal sequencing reads in a random set of cfDNA samples from patients with HCC and healthy individuals. The boxed regions are examples where patients with HCC and healthy individuals show differences in read distribution overlapped with histone marks or predicted enhancers. The red asterisk represents a predicted enhancer region from the ENCODE Project. Genomic positions are based on the human genome reference (hg19). (A) ESRRG (Chromosome 1q41; boxed region: chr1:216 700 111–216 704 999); and (B) SOX9 (Chromosome 17q24.3; boxed region: chr17:70 121 229–70 122 119). cfDNA, cell-free DNA; HCC, hepatocellular carcinoma; ENCODE, Encyclopaedia of DNA Elements.