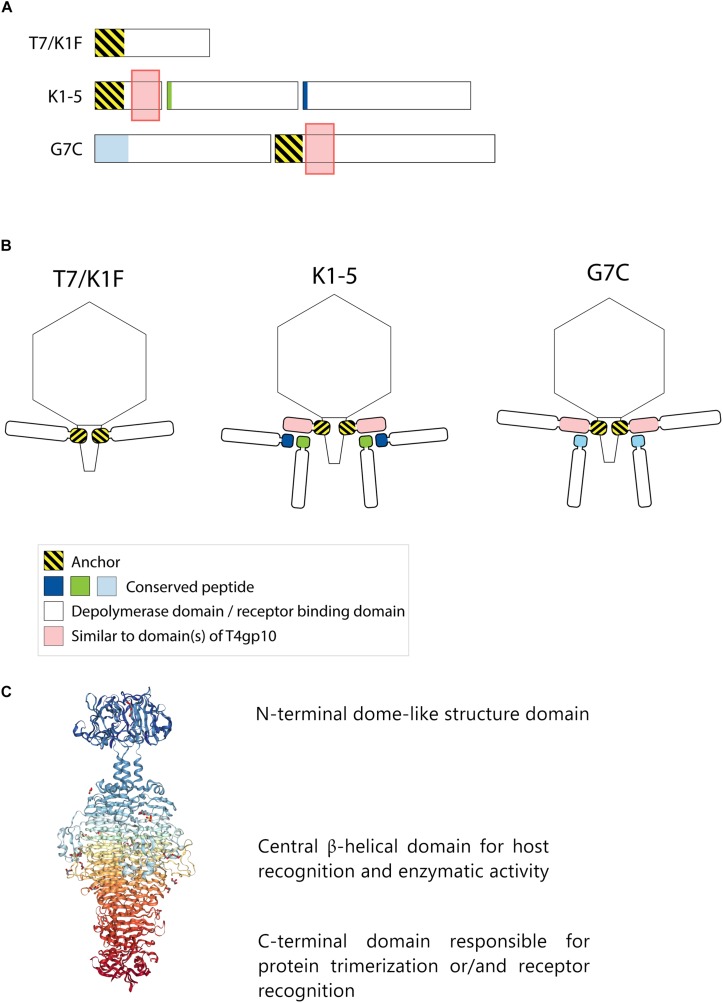
FIGURE 1.
Anchor and anchor-branched receptor binding protein (RBP) complexes confirmed by structural experiments. (A) The modular genetic organization of RBPs in single (T7 and K1F) and double RBP systems (K1-5 and G7C phages). (B) Schematic modeling of four different RBP systems in the virion structure. The T7 tail fiber (gene 17, T7p52) and K1F tail fiber (gene 17, CKV1F_gp36) have only an N-terminal anchor domain; K1-5 uses an adapter protein (gp37 with T4gp10-like domain) interacting with K5 lyase (gp46) and K1 endosialidase (gp47) via a conserved hepta- and undecapeptide, respectively; Phage G7C produces an anchor-branched complex with one anchored RBP (gp66) having a T4gp10-like domain and the second RBP connected via a conserved peptide to theT4gp10-like domain. (C) Modular structure of the model tail spike of Salmonella phage P22 (PDB ID 2XC1), illustrating a typical modular structure of RBPs. A N-terminal dome-like structure domain, a central β-helical domain for host recognition and enzymatic activity and a C-terminal domain responsible for protein trimerization or/and receptor recognition are shown (Berman et al., 2000; Seul et al., 2014; Rose et al., 2018).