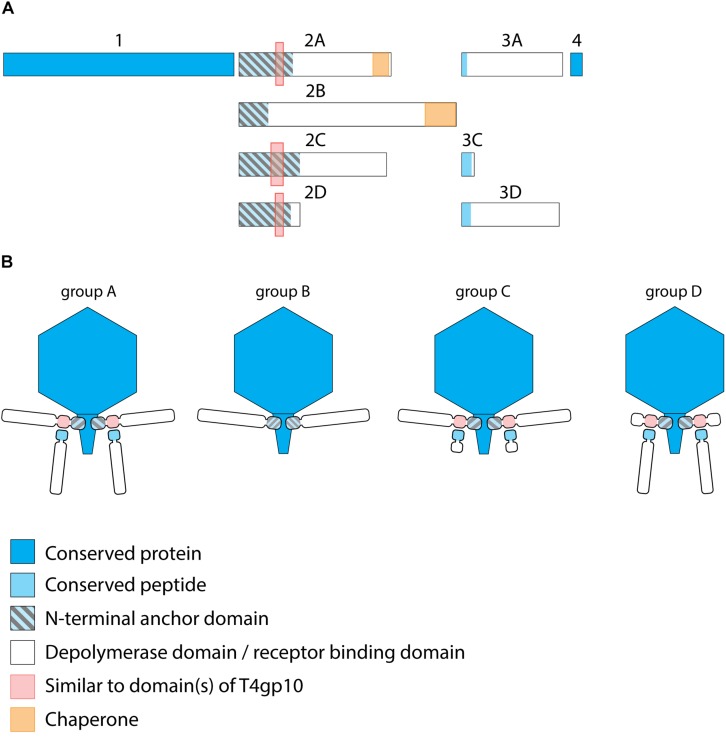
FIGURE 2.
Receptor binding protein systems of KP32viruses. Phages and their RBPs that are proposed to follow these systems, including their grouping into groups A, B, C and D, are summarized in Table 1. (A) The modular composition of RBP genes of phages belonging to four different groups (A, B, C, and D) is shown relative to the broken gene synteny of phage KP32. For simplicity, only one flanking gene conserved across all groups is shown at each side. Annotations are given according to GenBank or according to their modeled function in this study (between brackets): Protein (1) – internal virion protein D; (2) – tail fiber protein (2A,B,C – anchor with depolymerase; 2D – anchor with truncated protein); (3) – hypothetical protein (3A,D – depolymerase with conserved peptide, 3C – truncated protein with conserved peptide); (4) – lysis protein. (B) Schematic models of RBP systems in phage particles of KP32viruses. Group A – two RBPs: anchor-branch attachment mode; Group B – one RBP: anchor attachment mode; Group C – two RBPs: anchor-branch attachment mode, second RBP truncated; Group D – two RBPs: anchor-branch attachment mode, first RBP truncated.