Fig. 1.
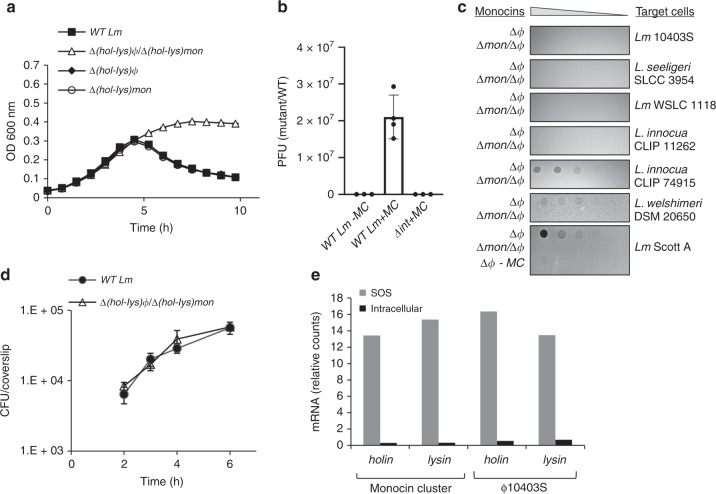
The Lm 10403S strain harbors two functional lytic phage elements. a Growth analysis of WT Lm and mutants harboring deletions of the elements lysis modules; LMRG_01552–4 of ϕ10403S (Δ(hol-lys)ϕ), and LMRG_02377-8 of monocin cluster (Δ(hol-lys)mon) or a mutant deleted of both lysis modules (Δhol-lys)ϕ/Δ(hol-lys)mon), in the presence of mitomycin C (MC). Growth analysis of the mutants without MC is presented in Supplementary Fig. 1. Error bars represent the standard deviation of three independent biological repeats, and are hidden by the symbols. b Plaque forming assay of WT Lm and a mutant deleted for the ϕ10403S integrase gene, Δint (LMRG_01511) with MC treatment (+MC). Virions obtained from MC treated bacterial cultures (4 h post MC treatment) or from bacteria grown to exponential phase (3 h, OD600 0.5) without MC treatment (WT Lm -MC), tested on an indicator strain for plaque forming units (PFU). Error bars represent the standard deviation of three independent experiments. c A monocin killing assay performed on monocins obtained from MC treated bacterial cultures of the Lm ϕ10403S-phage cured strain (Δϕ) and a mutant lacking both the monocin cluster (LMRG_02362-02378) and ϕ10403S-phage (Δmon/Δϕ), as a control. Five-fold serial dilutions of filtered supernatants (containing monocins) were applied on a lawn of different Listeria strains (target cells), and incubated for 1–2 days. The dark zones of growth inhibition indicate monocin killing activity. The experiment was performed three times. Monocins from Δϕ bacteria that were not treated with MC (Δϕ –MC) are shown at the bottom, as a reference. d Intracellular growth analysis of WT Lm and a deletion mutant lacking both lysis modules (Δ(hol-lys)ϕ/Δ(hol-lys)mon) in bone marrow derived macrophage (BMDM) cells. Growth curves represent one biological replicate, more independent experiments are shown in the source data file. Error bars represent standard deviation of a technical triplicate, sometimes hidden by the symbols. e Transcription analysis of the two phage elements holin and endolysin genes under SOS and intracellular growth conditions using NanoString technology (6 h post BMDM infection). Transcription levels are presented as fold change of relative counts for the indicated gene mRNA, relative to the levels observed during lysogeny (i.e., exponentially grown bacteria in BHI medium). Data represent 3 independent experiments. Source data are provided as a Source Data file.