Fig. 3.
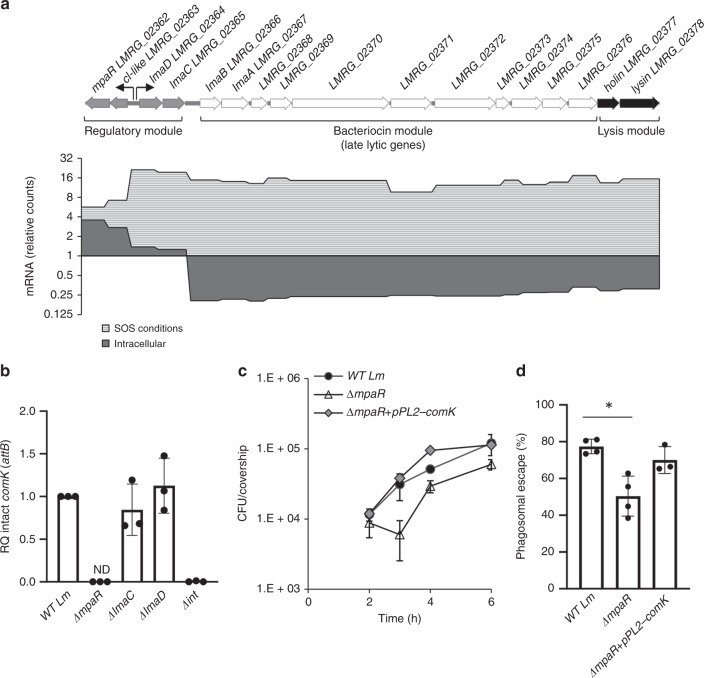
MpaR is required for ϕ10403S excision. a Upper panel: schematic representation of the monocin locus. The mon consists of 17 genes comprising: a regulatory module, a bacteriocin module encoding tail-like structures, and a lysis module. Lower panel: transcription analysis of the monocin cluster genes under SOS and intracellular growth (in BMDM cells) conditions, using NanoString technology. Transcription levels are presented as relative counts, relative to the levels observed at the lysogenic state (i.e., exponentially grown bacteria in BHI medium at 37˚C). Data represent three independent experiments. b qRT-PCR analysis of intact comK gene (representing ϕ10403S attB site) in WT Lm and indicated mutants grown intracellularly in BMDM cells (6 h post infection). Presented as relative quantity (RQ), relative to the levels in WT bacteria. The data is a mean of three independent experiments. Error bars indicate a standard deviation. c Intracellular growth analysis of WT Lm, ΔmpaR and ΔmpaR mutant complemented with an intact comK gene under its native promoter (ΔmpaR+pPL2-comK). Growth curves represent one biological replicate, more independent experiments are shown in Supplementary Fig. 2 and in the source file. Error bars represent standard deviation of triplicate samples, sometimes are hidden by the symbols. d A bacterial phagosomal escape assay. Percentage of bacteria that escaped the macrophage phagosomes at 2.5 h post infection, as determined by a microscope fluorescence assay. Macrophages were infected with WT Lm, ΔmpaR and ΔmpaR mutant complemented with an intact comK gene on the pPL2 plasmid (ΔmpaR+pPL2-comK). The data is a mean of three independent experiments. The error bar represent standard deviation. Asterisk (*) indicates statistical significance of p < 0.01 calculated using Student's t-test. Source data are provided as a Source Data file.