Fig. 7.
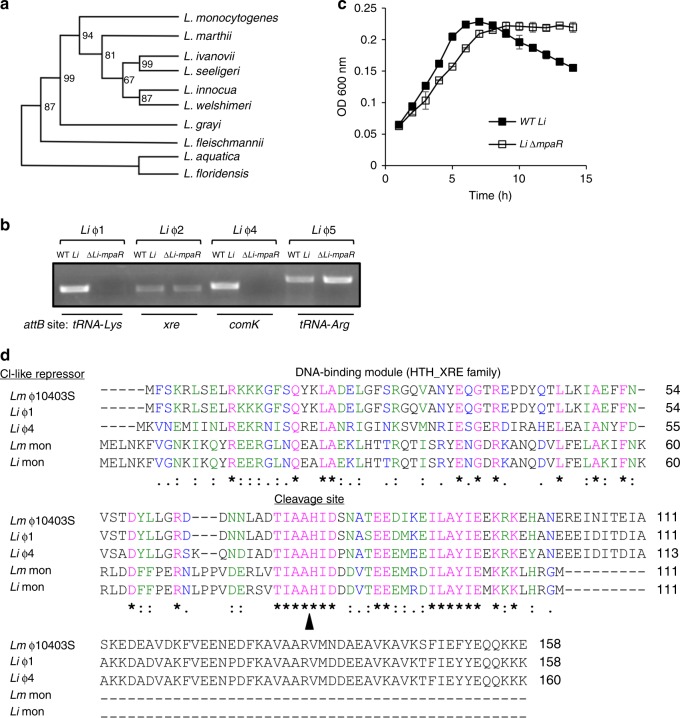
MpaR functions as a multi-phage coordinator. a Phylogenetic tree of Listeria species based on the amino acid sequence of MpaR (using 12 selected Listeria strains that represent the different Listeria species, as performed by Orsi et al. 2016). Bootstrap values are indicated on the relevant branches. b PCR analysis of the genomic region around the attB sites of L. innocua Clip112624 prophages, performed in wild-type bacteria (WT Li) and in a strain deleted of the mpaR gene (∆Li-mpaR) under MC treatment. Visible PCR products indicate phage excision and restoration of the attB sites. The genes harboring the prophages attB sites are indicated at the bottom. c Growth analysis of WT L. innocua (WT Li) and its isogenic mpaR mutant (Li-ΔmpaR) with MC (growth without MC is presented in Supplementary Fig. 6). The data shows the mean and the standard deviation of three independent biological repeats. Source data are provided as a Source Data file. d Multiple amino acid sequence alignment (by Clustal Omega) of the five different CI-like repressors that were found to be regulated by MpaR in Lm 10403S and L. innocua Clip112624 (Li). Identical amino acid residues (aa) are marked with (*), strongly similar aa are marked with (:), and weakly similar aa are marked with (.). The exact cleavage site of MpaR, as obtained by LC-MS/MS analysis of ϕ10403S CI-like repressor cleavage product, is indicated by a black arrowhead.