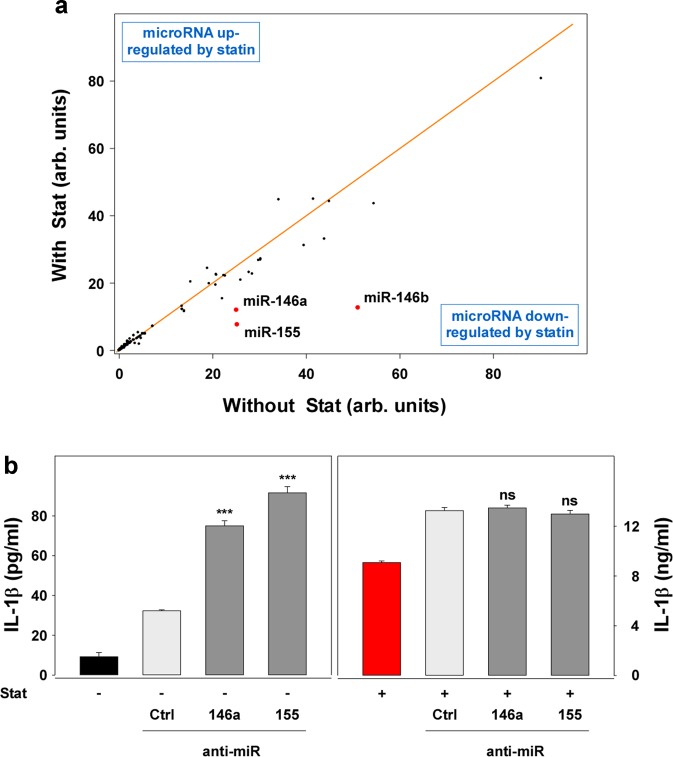
Fig. 5. microRNAs 146a, 146b, and 155 are down-regulated in statin-treated Mac.
a microRNA array. Total RNA was isolated from macrophages differentiated in the absence or presence of statin (25 cm2 flasks, 100,000 cells/cm2) with the “RNeasy Plus Mini Kit”. Deep sequencing was performed by the “Core Unit DNA”, Universität Leipzig, using the “TruSeq™Small RNA sample prepkit v2” (illumina, San Diego, USA). The mean of the normalized data of macrophages pretreated without statin and macrophages pretreated with statin was calculated and data of samples with a mean >100 counts (192 samples; compare Supplementary Table 2) were included into the analysis and blotted against each other. The orange line indicates unchanged expression. The red dots mark three selected miRs, which were down-regulated in statin-pretreated macrophages, as compared to Mac prepared in the absence of statin (compare Supplementary Table 2). A second array showed a similar result. b Blockade of miR-146a and miR-155 reverses the hypo-responsiveness in macrophages only to some degree. Macrophages were incubated as described in Fig. 1a (6-well plate; 100,000 cells/cm2). The respective “miRCURY LNA™” anti-miR (Exiqon, Qiagen, Vedbaek, Denmark) were prepared in “Lipofectamine RNAiMax” and 250 µl of this solution was added to 2750 µl of culture medium in the absence (−) or presence (+) of statin. After 24 h LPS was added. After further 24 h, the supernatants were harvested and analyzed in ELISA. Four experiments with similar results were performed. Data analysis and color code as in Fig. 1c (“Ctrl” vs. “anti-miR”).