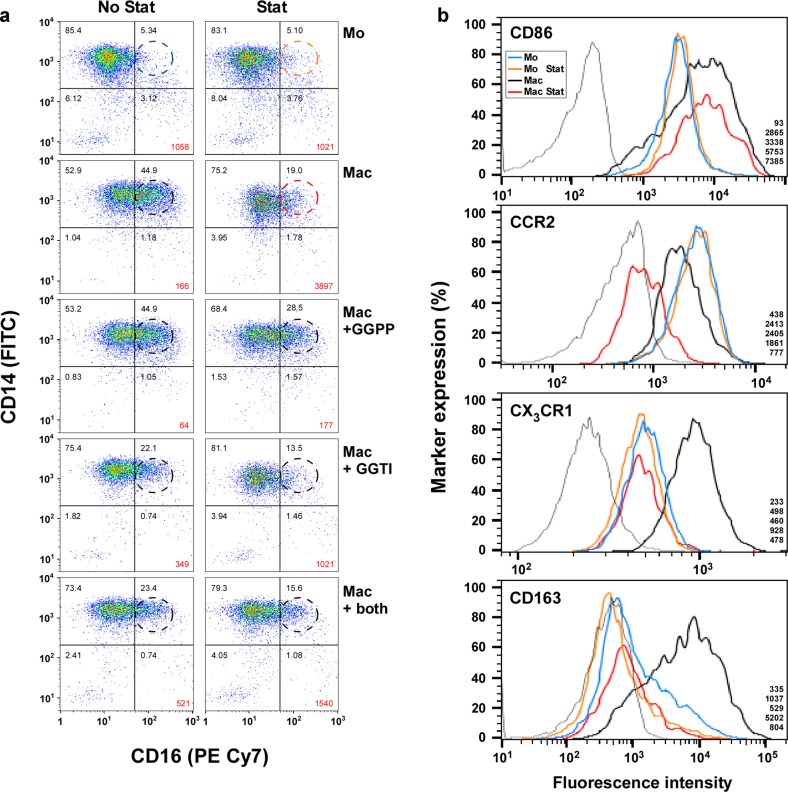
Fig. 6. The retainment effect is paralleled by reciprocal expression of macrophage-related surface markers.
a The enhanced CD14/CD16-expression in macrophages prepared without statin is not observed in macrophages prepared in the presence of statin. The surface marker expression of LPS-stimulated Mo and Mac prepared as described in Fig. 1a was analyzed by flow cytometry. For this purpose the cells (25 cm2; 100,000/cm2) were harvested, centrifuged (300 × g; 10 min), resuspended in 100 µl PBS and transferred into conical 96-well plates (Greiner). The plates were centrifuged (400 × g; 3 min) and incubated in PBS containing Zombie Aqua™ (BioLegend, San Diego, USA) for 15 min, followed by centrifugation and resuspension in PBS containing 1% BSA, 0.1% sodium azide and 1 mM EDTA (FACS-buffer). In order to avoid unspecific binding of the antibodies, the cells were incubated with 10% FcR-blocking reagent (Miltenyi) for 15 min at 4 °C in FACS-buffer. Antibodies against CD14 or CD16 (compare Supplementary Table 1) were added (15 min; 4 °C; in the dark). Analysis was performed in a LSR-Fortessa™, using the “FlowJo LLC” software (Ashland, OR, USA). Aggregated cells were excluded by FSC-H- and FSC-A-scatter and dead cells were excluded by gating Zombie Aqua™-negative cells (compare “gating strategy” in Supplementary Fig. 5A–D). The dashed circles indicate the same position of CD14+/CD16+-cells in each graph. Five experiments with similar results were performed. The numbers in the gates reflect the respective percentages. B The CD163- and CX3CR1-expression is upregulated in untreated macrophages, but not in statin-treated macrophages. Mo and Mac were prepared as described in Fig. 1a in 25 cm2 flasks (57,353 cells/cm2). After the respective incubation, the cultures were gently scraped, the cells centrifuged (300 × g; 10 min), the supernatants harvested and the cell pellets resuspended twice in 1 ml MACS-buffer (PBS, 2 mM EDTA, 2% FCS; 4 °C). FcR-blocking reagent (2%; Miltenyi) was added for 10 min. Antibodies against CD86, CCR2/CD192, CX3CR1 or CD163, or the respective isotype controls (compare Supplementary Table 1; thin gray line in the figure) were added. After 20 min of incubation in the dark, the cells were washed with MACS-buffer and analyzed using a LSR-Fortessa™ (BD Biosciences). Aggregated cells were identified in the FSC-H (forward scatter-high) FSC-A (forward scatter-area) window and excluded from the analysis (compare “gating strategy” in Supplementary Fig. 5A–D). The monocyte region was then determined and gated based on the FSC and SSC (side scatter) parameters. Dead cells were excluded by gating of cells, which were not stained for 7-AAD (7-aminoactinomycin D; BD Biosciences). Visualization and analysis were performed using the “FlowJo LLC” software and the expression of the respective marker (normalized to ratio) was presented. A representative experiment out of seven is shown. The numbers in the lower right corner reflect the MFI (geometric mean) of “isotype control”, “Mo”, “Mo + Stat”, “Mac,” and “Mac + Stat”, respectively, taken from “FlowJo LLC”.