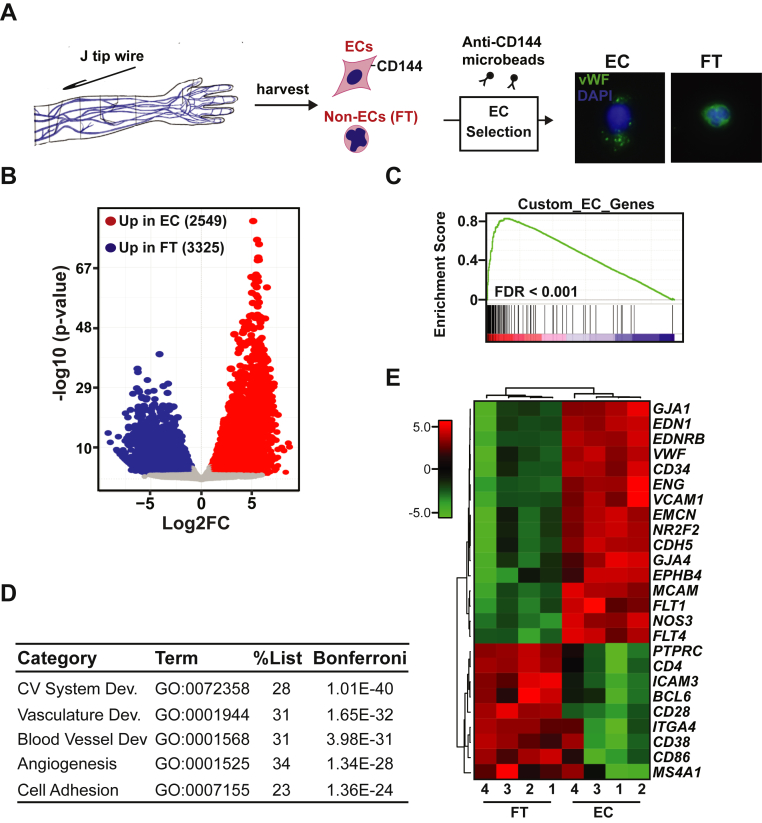
Figure 1.
Ex Vivo CD144-Selection Enriches Vascular ECs in Patient-Derived Samples
(A) Schematic of endothelial cell (EC) isolation procedure, selection of ECs using anti-CD144 magnetic microbeads and photomicrographs of immunofluorescence for von Willebrand factor (VWF) (green) in CD144 selected ECs and flow through cells (FT = nonselected). Nuclei were counterstained with DAPI (blue). Results shown at 40x magnification. (B) Volcano plot showing Log2 fold change (Log2FC) expression vs. log10 (p value) of differentially expressed genes in CD144-selected cells vs. FT. For significance cutoff, false discovery rate (FDR) adjusted p value <0.05 and 2-fold change was used. (C) Gene set enrichment plot showing positive enrichment of genes expressed in CD144-selected cells in a curated EC gene set. (D) Table showing results of gene ontology analysis (ToppGene suite) of differentially expressed genes from CD144-selected cells vs. FT cells. Bonferroni adjusted p value is shown. All statistically significantly upregulated genes in CD144 selected cells vs. FT cells from Figure 1B were included in the analysis using a cutoff of 2-fold; FDR <0.05. GO: Biological Processes are listed. (E) Heat map showing expression data of specific EC-restricted and leukocyte-restricted marker genes in EC and FT samples. Samples were clustered using Pearson correlation. For each EC and FT patient-derived sample, pairwise analysis was performed. Numbers below the heat map indicate paired samples.