Figure 2.
Comparative Transcriptomics Identifies Changes in Pathways of Proliferation, Metabolism, and Androgen Signaling in ECs From Patients With T2DM
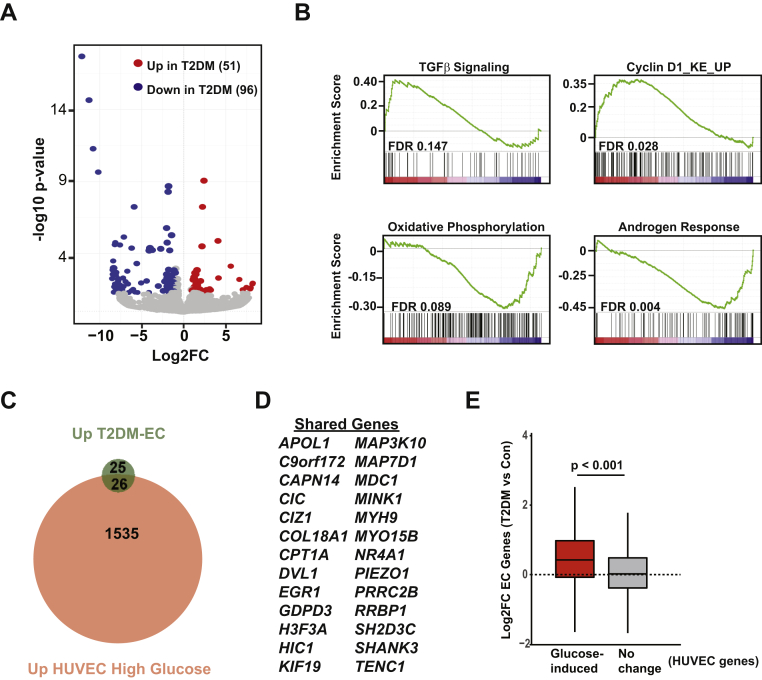
(A) Volcano plot showing Log2 fold change (Log2FC) in expression of protein coding genes vs. log10 (p value) in T2DM (endothelial cells) ECs compared with EC-Con (Same RNA-seq data from Figure 1). False discovery rate (FDR adjusted p value cutoff <0.05. (B) Gene set enrichment plots in T2DM ECs vs. EC-Con. Genes were ranked using the “stat” parameter generated in the DESeq2 pipeline. For each plot, FDR is included. (C) Venn diagram showing overlap in genes induced in T2DM-ECs (green) and HUVEC treated with 25 mM glucose for 24 h (red). (D) List of shared genes from panel C. (E) Boxplot showing median Log2FC (T2DM ECs vs. EC-Con) of all HUVEC genes induced by high glucose (2-fold, FDR <0.05) or HUVEC genes that did not change with glucose stimulation. For E, p value was determined by Wilcoxon rank sum test.