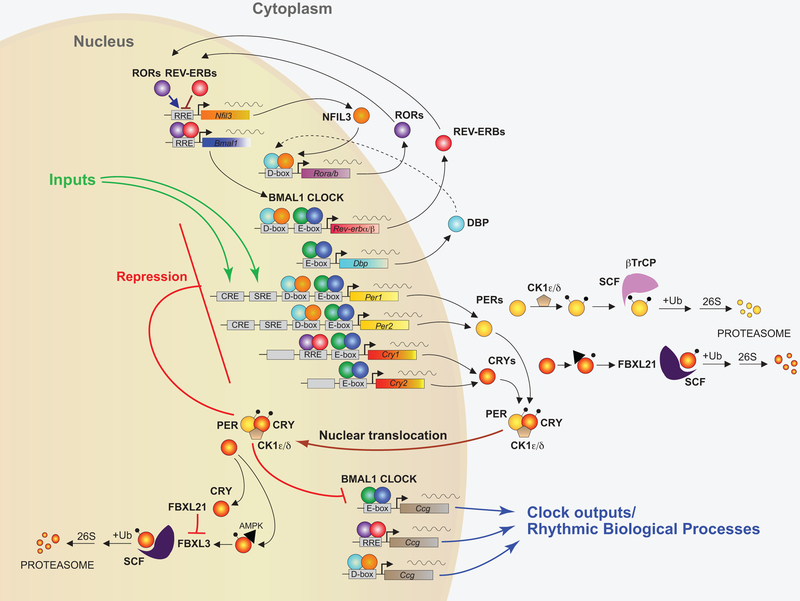
Figure 1: Core components of the mammalian circadian clock.
In the core feedback loop, the transcription factors BMAL1 (green circles) and CLOCK (blue circles) bind to E-box domains on gene promoters, including the genes for Per1 and Per2 (yellow) and Cry1 and Cry2 (red/yellow). PERs (yellow circles) and CRYs (red/yellow circles) dimerize and translocate to the nucleus after binding with casein kinase δ (CK1δ) or CK1ϵ, where they repress their own transcription. The stability of PER and CRY is regulated both in the cytoplasm and within the nucleus by several proteins, including FBXL21 and FBXL3. In a second feedback loop, CLOCK and BMAL1 also regulate the transcription of genes for the nuclear receptors REV-ERBα and REV-ERBβ (red circles), which compete with the retinoic acid-related orphan receptors, RORα, RORβ, and RORγ (purple circles) for binding to RRE elements on the BMAL1 gene promoter, providing both positive (ROR) and negative (REV-ERB) regulation of BMAL1 transcription. A third feedback loop is mediated by CLOCK/BMAL1-mediated transcription of the gene Dbp (light blue) and the ROR/REV-ERB-mediated transcription of Nfil3 (orange). DBP (light blue circles) and NFIL3 (orange circles) dimerize and bind to D-box elements on the promoters of many of the core clock genes, providing additional layers of regulation. In addition, CLOCK/BMAL1, ROR/REV-ERB, and DBP/NFIL3 regulate the transcription of many other clock output genes (Figure modified from (Takahashi, 2017)).