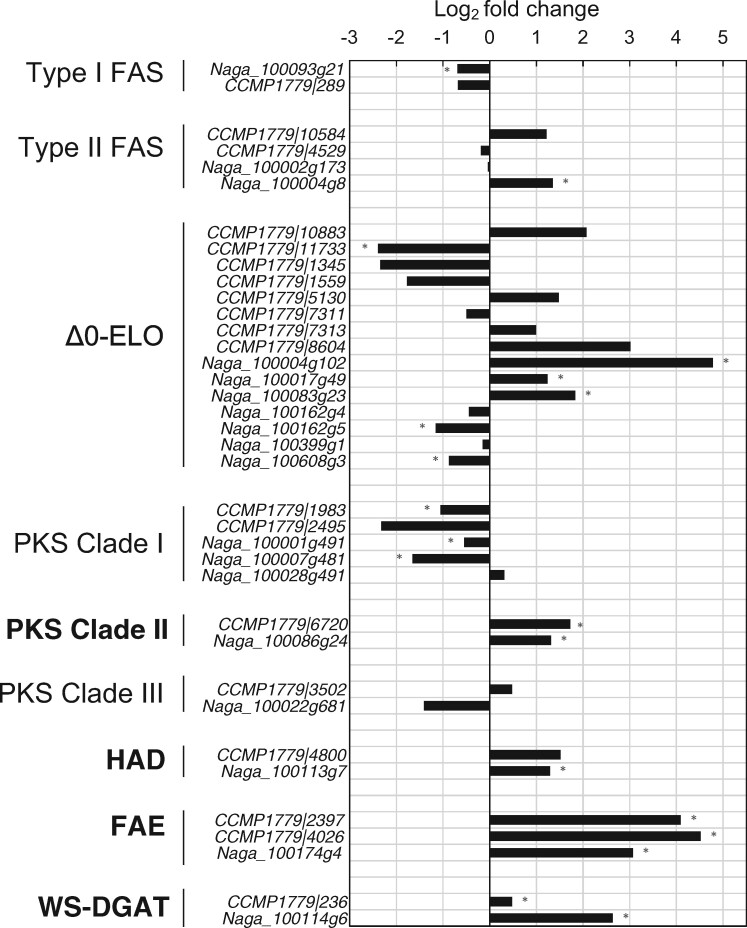
Fig. 3.
Expression level of the genes potentially coding the enzymes catalyzing the different reactions involved in the biosynthesis of saturated C14–18 fatty acids, LCHFAs, LCAs, and LCDs as well as the formation of ester bonds within the cell wall biopolymer. The enzymes most likely involved in processing fatty acids ≥C18 and coded by genes upregulated under dark conditions are in bold. The horizontal axis indicates the log2 fold change in gene expression, between the dark treatment and the two control treatments (Control 1 and Control 2). Significant differences (P < 0.01) are indicated with an asterisk. The prefixes Naga and CCMP1779 denote transcripts from N. gaditana CCMP526 and N. oceanica CCMP1779, respectively. FAS, fatty acid synthase; Δ0-ELO, Saturated fatty acid elongases; PKS, polyketide synthase; HAD, hydroxyacyl-acyl carrier protein-dehydratase; FAE, fatty acid elongation enzyme; WS-DGAT, bifunctional wax ester synthase/diacylglycerol acyltransferase.