Abstract
The 57 kDa antigen recognized by the Ki-1 antibody, is also known as intracellular hyaluronic acid binding protein 4 and shares 40.7% identity and 67.4% similarity with serpin mRNA binding protein 1, which is also named CGI-55, or plasminogen activator inhibitor type-1-RNA binding protein-1, indicating that they might be paralog proteins, possibly with similar or redundant functions in human cells. Through the identification of their protein interactomes, both regulatory proteins have been functionally implicated in transcriptional regulation, mRNA metabolism, specifically RNA splicing, the regulation of mRNA stability, especially, in the context of the progesterone hormone response, and the DNA damage response. Both proteins also show a complex pattern of post-translational modifications, involving Ser/Thr phosphorylation, mainly through protein kinase C, arginine methylation and SUMOylation, suggesting that their functions and locations are highly regulated. Furthermore, they show a highly dynamic cellular localization pattern with localizations in both the cytoplasm and nucleus as well as punctuated localizations in both granular cytoplasmic protein bodies, upon stress, and nuclear splicing speckles. Several reports in the literature show altered expressions of both regulatory proteins in a series of cancers as well as mutations in their genes that may contribute to tumorigenesis. This review highlights important aspects of the structure, interactome, post-translational modifications, sub-cellular localization and function of both regulatory proteins and further discusses their possible functions and their potential as tumor markers in different cancer settings.
Keywords: Cancer, Cell signaling, Regulatory protein, Protein interactions, Post-translational modifications
Core tip: Intracellular hyaluronic acid binding protein 4 and serpin mRNA binding protein 1 are paralog human regulatory proteins that share 41% amino acid sequence identity. The characterization of their protein interactomes suggested their functional association with transcriptional regulation, mRNA metabolism and in the cell’s DNA damage and stress responses. Their complex post-translation modifications, involving phosphorylation, arginine methylation and SUMOylation, as well as their finely regulated sub-cellular localization in the nucleus and cytoplasm as well as in several cytoplasmic and nuclear granules suggest extensive functional regulation. This review discusses the functional and structural aspects and emerging roles of these regulatory proteins in human cancer.
INTRODUCTION
Ki-1 was the first monoclonal antibody specific for Hodgkin and Stenberg-Reed cells in Hodgkin’s lymphoma[1]. Ki-1 recognizes CD30, a glycoprotein of 120 kDa found on the surface of Hodgkin’s cells, and cross-reacts with an intracellular antigen of 57 kDa, named Ki-1/57, which was functionally and structurally uncharacterized at that time[2,3]. Ki-1/57 was also named intracellular hyaluronic acid binding protein 4 (IHABP4; GeneBank: AF241831) as it bound to hyaluronic acid in vitro[4]. Huang and co-workers also observed that IHABP4 binds to others negatively charged molecules such as glycosaminoglycans, e.g. chondroitin sulfate and heparin sulfate, and to RNA. However, the functional role of these interactions is not completely understood. The recommended name is hyaluronic acid binding protein 4 (HABP4).
HABP4 shares 40.7% identity and 67.4% similarity with serpin mRNA binding protein 1 (SERBP1), indicating that they might be paralogs, possibly with similar or redundant functions in human cells[5]. The name of its putative paralog was originally CGI-55, derived from: “Comparative Gene Identification”, a method used to search for related genes. In the year 2000, Lin’ s group obtained 150 potential full-length novel human genes through CGI, identified from the Caenorhabditis elegans proteome[6], with the number 55 being one of the SERBP1 transcript variants (GenBank: AF151813). Independently, CGI-55 was identified as an interactor of plasminogen activator inhibitor type-1 (PAI-1) RNA; therefore, it was also called PAI-1 RNA-binding protein or PAI-RBP1[7]. Moreover, other names such as HABP4L and SERPINE 1 were also used. As SERBP1 is most widely used it will be adopted in this review.
Since the identification of these two proteins, several studies have addressed their structure and function. Here, we present a detailed report on the current knowledge on the HABP4 and SERBP1 proteins.
HABP4 AND SERBP1 STRUCTURE
Structurally, HABP4’s amino acid sequence, has a high level of disorder-promoting amino acids (Alanine, Arginine, Glycine, Glutamine, Serine, Proline, Glutamic acid, Lysine), a high net charge and a low mean hydropathy value in its amino acid composition[8]. These features are observed for most intrinsically unstructured proteins (IUP) and inhibit the formation of a hydrophobic core or a regular secondary structure[9,10]. Bressan et al[8] demonstrated using size exclusion chromatography (SEC), analytical ultracentrifugation and small angle X-ray scattering (SAXS) studies on the HABP4 C-terminal region (HABP4122-413), that it is an elongated monomer in solution, without a well-defined core. Thus, the HABP4 C-terminal has been shown to be a pre-molten globule of 37 kDa.
A proteinase K sensitivity assay showed that HABP4122-413 was readily degraded, confirming its flexibility and absence of a stable hydrophobic core. Additionally, the spectrum obtained by circular dichroism (CD) experiments was typical of a random coil or denatured proteins, indicating the absence of a regular secondary structure[8]. However, after the addition of 2,2,2-trifluoroethanol (TFE), the CD pattern for HABP4 shifted, showing an increase in secondary structure. TFE is an alcohol used to promote increased hydrogen bonding, and thus increases propagation of the secondary structures in polypeptides[11,12]. The appearance of secondary structure is commonly seen for IUPs when they associate with their interactors[13]. The gain of the structure may be explained by the existence of secondary structural elements in the protein sequence[14].
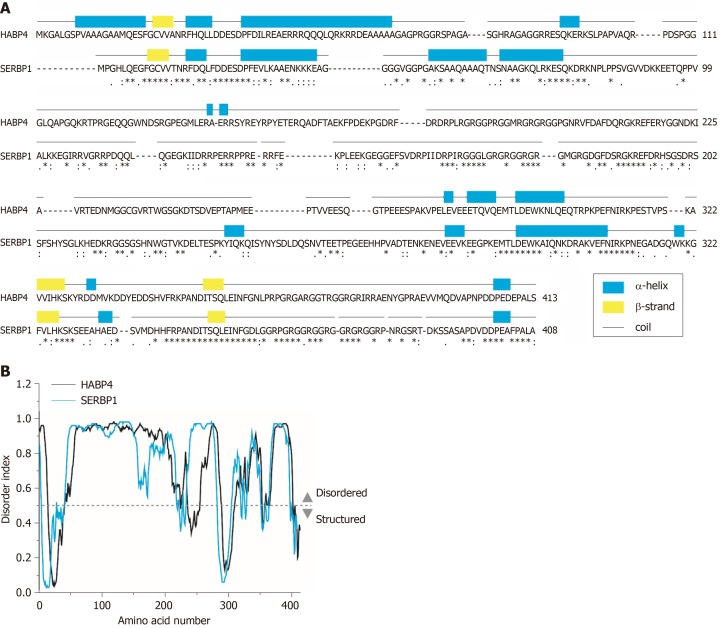
Despite the lack of structural studies for SERBP1, its high level of similarity and identity to HABP4 on the amino acid sequence level, allowed comparative bioinformatics analyses, which suggested that SERBP1 may also be an IUP. The protein secondary structure prediction (PSIPRED) analysis of both proteins illustrated that both HABP4 and SERBP1, have similar contents of predicted secondary structure and random coil. This may lead to the conclusion that that both HABP4 and SERBP1, are unstructured proteins (Figure 1).
Figure 1.
Bioinformatics analysis of hyaluronic acid binding protein 4 and serpin mRNA binding protein 1 amino acid sequences. A: Alignment between hyaluronic acid binding protein 4 (HABP4) and serpin mRNA binding protein 1 (SERBP1) and their predicted secondary structure content obtained by Clustal Omega and PSIPRED 4.0, respectively; B: Predictable disorder of HABP4 and SERBP1 structure obtained by DISOPRED 3. Below amino acids: Asterisk: Identical amino acid residues; colon: Strong similar properties; period: Weak similar properties. HABP4: Hyaluronic acid binding protein 4; SERBP1: Serpin mRNA binding protein 1.
Proteins belonging to the IUP family are associated with a plethora of cellular processes, such as translation, RNA recognition, transcriptional regulation, cell cycle control, membrane fusion and transport, protein phosphorylation, storage of small molecules and the regulation and assembly of protein complexes[13,15]. All these biological processes are in accordance with the present knowledge on the interaction network of HABP4 and SERBP1 (see the Functional aspects of HABP4 and SERBP1 in the following sections for more details).
POST-TRANSLATIONAL MODIFICATIONS
Post-translational modifications (PTMs) control protein functions by covalently attaching molecules to specific amino acid residues. The types of modifications exceed 200, such as phosphorylation, glycosylation, methylation, acetylation, ubiquitinylation, and SUMOylation among others, with phosphorylation being the most widely studied[16-19].
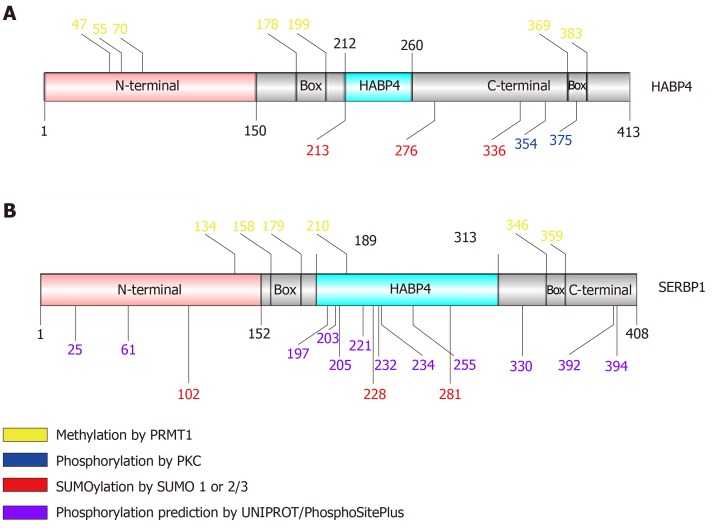
The PTMs of HABP4 and SERBP1 have been discovered over the years mainly due to the identification of their interaction with modifying proteins. Until now, HABP4 and SERBP1 were predicted to have phosphorylation, methylation and SUMOylation sites, as shown in Figure 2[20-22]. These PTMs and their impact on the functions of HABP4 and SERBP1 are described below.
Figure 2.
Schematic view of hyaluronic acid binding protein 4 and serpin mRNA binding protein 1 primary structure identifying residues that exhibit post-translational modification. The pink region corresponds to the N-terminal domain; gray corresponds to the C-terminal; light blue corresponds to the HABP4 domain. “Box” indicates RGG/RXR boxes.
Phosphorylation
Initially, immunoprecipitates of HABP4 from three different tumor cell lines (L540, U266/B1 and Raji Burkitt), using the Ki-1 antibody, revealed an associated serine/threonine protein kinase activity[23]. Based on this it was also hypothesized that HABP4 enzymatic activity could be regulated by self-phosphorylation. This hypothesis was ruled out by Nery and co-workers[20], who demonstrated that full-length recombinant HABP4 did neither exert kinase activity itself nor towards other proteins. These results are in agreement with the sequence analyses that also do not show any kinase domain features in the HABP4 amino acid sequence (Figure 2).
Yeast two-hybrid (Y2H) screens identified the Receptor of ACtivated Kinase 1 (RACK1), a protein kinase C (PKC) adaptor protein, as a HABP4 interactor and it was hypothesized that HABP4 could be a substrate for phosphorylation by PKC, explaining the earlier findings of the co-precipitated kinase activity[23].
Experiments with L540 cells showed that HABP4 is indeed phosphorylated by PKC and that its phosphorylation level is increased when cells are activated by the addition of phorbol 12-myristate 13-acetate (PMA)[20]. Furthermore, HABP4 can be phosphorylated by different PKC isoforms, such as PKCαβ, PKCδ, PKCλ/ζ and more strongly by PKCθ. PKCµ was the only member of the PKC family that did not phosphorylate HABP4 in vitro[20].
The phosphorylation of HABP4 does not seem to be affected by the presence or absence of RACK1, which binds to HABP4 C-terminal domain. On the other hand, the interactions between HABP4 and nuclear proteins, including RACK1 and CHD3[5,20], were down-regulated in response to phosphorylation. This modulation of interaction in response to phosphorylation shows that HABP4, at some level, may regulate the functions of the adaptor protein RACK1 or other protein interactors.
PKC phosphorylates HABP4 only in its C-terminal domain, which contains 15 potential Ser/Thr residues that could be targets of phosphorylation. Interestingly, only two of the threonine residues (T354 and T375) were indeed phosphorylated in vitro by commercial PKC-Pan, showing that PKC activity on HABP4 is highly specific[20] (Figures 2A and 3A).
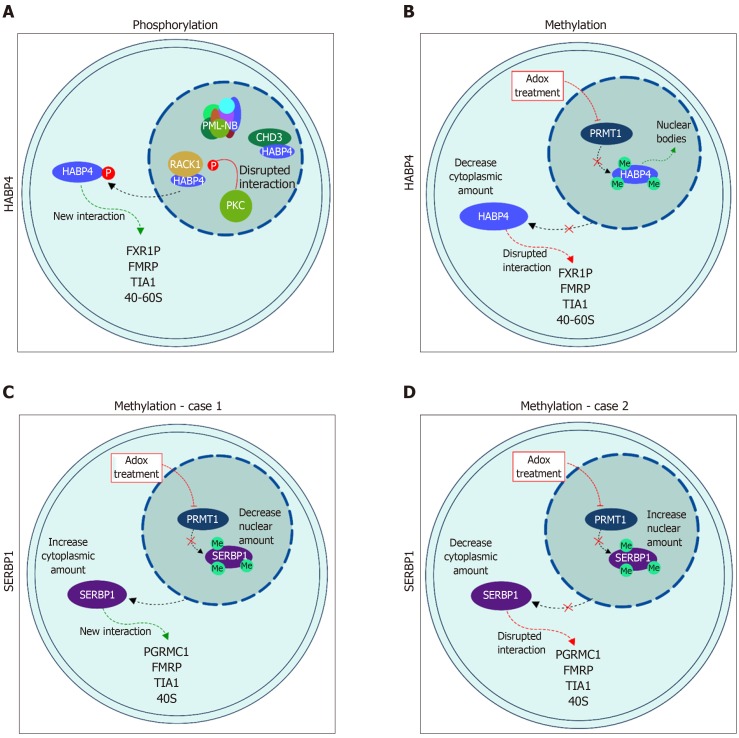
Figure 3.
Cellular localization of hyaluronic acid binding protein 4 and serpin mRNA binding protein 1 in response to post-translational modification. A: Localization of hyaluronic acid binding protein 4 (HABP4), in light blue, after phosphorylation; and B: after methylation; C and D: Localization of serpin mRNA binding protein 1 (SERBP1), in purple, after methylation. SERBP1 methylation is still controversial and both possibilities are presented as related in the literature: C[26] and D[28]. Please see text for details. HABP4: Hyaluronic acid binding protein 4; SERBP1: Serpin mRNA binding protein 1.
More recently, SERBP1 was also established as a RACK1 interactor through Y2H screens[24]. Since RACK1 can recruit PKC, we can predict that SERBP1 is also likely to be modified by phosphorylation, since several of the Ser/Thr residues are conserved in both amino acid sequences (Figure 2).
Methylation
Many of the cellular processes that are mediated by specific interactions between proteins and other proteins or nucleic acids, are regulated by arginine methylation. The RGG/RXR box, where X is any amino acid, is the main target of arginine methyltransferases, such as PRMT1. In general, these motifs are found in proteins related to transcriptional regulation and RNA processing. HABP4 and SERBP1 present conserved RGG/RXR boxes in their sequences, localized mostly in their C-terminal regions (Figure 2). Both, HABP4 and SERBP1, were methylated by PRMT1 in vitro[26-28]. In vivo assays with L540 cells showed that the levels of HABP4 decrease in the cytoplasm in response to the methylation inhibitor Adox (adenosine-2',3'-dialdehyde) treatment, indicating that the methylation status of HABP4 can affect its cellular distribution. Additionally, nuclear HABP4 was stronger methylated than that localized in the cytoplasm. Interestingly, the paralog SERBP1 behaved otherwise. In untreated cells, SERBP1 in mostly found in the nucleus and upon Adox treatment more in the cytoplasm[26] (Figure 3C).
The interaction between HABP4 and RACK1 did not influence the methylation pattern. Although RACK1 interacts with the C-terminal domain of HABP4, methylation of the RGG/RXR box cluster 369-383 continued to occur. However, it was reported that significant inhibition of methylation in the C-terminal domains of HABP4 and SERBP1 occurred, when they were previously phosphorylated[26-28].
SUMOylation
SUMOylation is the attachment of Small Ubiquitin-like Modifier (SUMO) proteins to a lysine residue in the specific target protein. This PTM regulates proliferation[29], transcription[30], mRNA processing and metabolism[31], among many other processes. The insight that HABP4 and its paralog SERBP1 could be SUMOylated was derived by the finding that both interact with proteins related to the SUMOylation machinery, including: UBC9, PIAS3 and TOPORS interacting with HABP4 and UBA2, PIAS-1/ -3 /-y and TOPORS all interacting with SERBP1[32,33].
HABP4 has seven predicted sites for SUMOylation, of which three are highly likely: Lysine 213, 276 and 336 (Figure 2A). Likewise, SERBP1 exhibited fifteen potential SUMOylation sites, of which six had a higher probability of being conjugated with SUMO (Figure 2B). In vitro assays showed that HABP4 is indeed SUMOylated by SUMO-2/3 at the three main targets[22]. In vivo experiments revealed that wild-type HABP4, but not the SUMOylation-defective mutant HABP4K213R/K276R/K336R, co-immunoprecipitated with anti-SUMO-1 and anti-SUMO-2 antibodies.
The same approach was used for SERBP1, and mutations in the three lysine residues with the highest score (K102R/K228R/K281R), resulted in SERBP1 being unavailable for modification by SUMO conjugation that was observed in the wild-type protein[22].
SUMOylation by SUMO-1 or SUMO-2 does not affect the profile of SERBP1’s interaction partners. The analysis of the partners identified by immunoprecipitation followed by tandem mass spectrometry (IP-MS/MS) with or without SUMO-1 or SUMO-2 transfection, resulted in the identification of proteins related to gene expression regulation. Specifically proteins involved in transcriptional control, RNA splicing and translation, ribosome biogenesis, apoptosis or mitosis, but no significant differences were observed between SUMO-1 and SUMO-2 co-transfection[22].
On the other hand, HABP4 displayed functional differences when co-expressed with SUMO-1 or SUMO-2. HABP4 co-immunoprecipitated 68 proteins when co-expressed with SUMO-1, whereas only 29 proteins were detected when HABP4 was co-expressed with SUMO-2. The enrichment of biological processes also presented some differences: HABP4 co-expressed with SUMO-1 was found to be involved in the regulation of transcription, RNA splicing, translation, ribosome biogenesis, mitotic cell cycle, the apoptotic process, and DNA repair. However, when HABP4 was co-expressed with SUMO-2, much fewer biological processes were observed, such as gene expression regulation (transcription, RNA splicing and translation) and telomere maintenance[22]. In summary, these data showed that HABP4 and SERBP1, despite having similar modification sites, respond in different ways to these PTMs in the context of their protein interactomes.
Cellular localization of HABP4 and SERBP1
More than two decades ago, Rhode and co-workers showed that HABP4 has both cytoplasmic and nuclear localizations. In the nucleus it is often associated with heterochromatin, euchromatin, and the nucleolus[3]. The paralog SERBP1 also exhibited shuttling between cytoplasm and nuclei[5]. Since then, numerous data on these protein localizations have been reported. For example, Nery and co-workers[20] showed that the phosphorylation of HABP4 by PKC affects its cellular localization. Upon PMA-stimulus, HABP4 was no longer found in the nucleus, whereas its cytoplasmic level increased (Figure 3A).
Methylation also influences HABP4 and SERBP1 localization patterns. The methylation inhibition by Adox treatment leads to a decrease in HABP4 cytoplasmic staining, but shuttling from the nucleus to the cytoplasm of SERBP1[26]. The localization of SERBP1 is still controversial. Lee and co-workers[27,28], showed that SERBP1 nuclear staining is stronger than cytoplasmic staining after methylation inhibition (Figure 3D). Altogether, these data suggest that HABP4 and SERBP1 are involved in nuclear functions, this possibly depends on the phase of the cell cycle, or on specific cell growth conditions.
Once in the nucleus, HABP4 was detected at sub-structures such as nucleoli, where ribosome biogenesis and maturation occur[34]. Both, SERBP1 and HABP4, were found to co-localize with p80-coilin, a marker for Cajal bodies, and HABP4 has also been observed to co-localize with GEMS (Gemini of coiled bodies) in cells treated with Adox[33,34]. Cajal bodies and GEMS are both considered nuclear compartments involved in small nuclear ribonucleoproteins snRNP storage or the assembly of pre-mRNA splicing complexes[35,36]. Upon Adox treatment, HABP4 has also been shown to co-localize with SC-35, a marker protein for splicing speckles[34]. These sub-compartments are known to store pre-mRNA splicing complexes, and HABP4 interacts with SFRRS1/9 and hnRNPQ, both known as splicing regulatory proteins[34]. In summary, these data suggest that methylation not only promotes nuclear import but also directs HABP4 more specifically to selected nuclear bodies.
The fact that HABP4 and SERBP1 interacted with several proteins related to promyelocytic leukemia nuclear bodies (PML-NBs), raised interest to determine if both proteins would play a role in the formation and distribution of these bodies. PML-NBs are related to protein modification, transcriptional regulation, DNA-damage response, DNA repair, cell proliferation and apoptosis[37]. The relationship of HABP4 and SERBP1 with PML-NBs was explored by Saito and co-workers, who showed that the number of PML-NBs decreases in response to HABP4 or SERBP1 over-expression. After treatment with arsenic trioxide (As2O3), which under normal conditions increased PML-NBs formation, their abundance was significantly lower in cells over-expressing HABP4 and SERBP1. Interestingly, SERBP1 and HABP4 SUMOylation-defective mutants revealed distinct behaviors in relation to PML-NBs. While the SERBP1 SUMOylation-defective mutant displayed an effect similar to the wild-type protein, the HABP4 SUMOylation-defective mutant did not have any impact on the number of PML-NBs. In addition, the presence of HABP4 or SERBP1 seems to affect the diffuse distribution of the PML protein[22].
The regulation of HABP4´s localization to PML-NBs by SUMOylation is exactly the opposite of what was observed for PML protein. PML only co-localizes with nuclear bodies when it is SUMOylated[38]. Although HABP4 and SERBP1 influenced the formation and distribution of PML-NBs, the PML protein, the main component of the PML-NBs, did not interact with HABP4 and SERBP1 in IP-MS/MS experiments. It is known that changes in the formation and distribution of PML-NBs can also occur in response to UV-C light exposure in a p53-dependent manner[39]. It has also been well established that HABP4 interacts with p53, reducing its transcriptional activity[32]. It could be hypothesized that the role of HABP4 in PML-NBs formation is mediated by regulation through p53, which on the other hand is part of PML-NBs.
Outside the nucleus, HABP4 interacts with the Fragile X-Related Protein 1 (FXR1P) and the Fragile X Mental Retardation Protein (FMRP), both of which are related to translational regulation[40,41]. Furthermore, it co-localizes with TIA1 upon arsenite challenge, which is a marker protein for stress granules[40,42].
In ribosomal fractions, HABP4 was detected in fractions containing the 40 and 60S ribosomal subunits and in small amounts in the polysome fraction, thereby pointing to possible additional roles of HABP4 in the context of translation[40]. Furthermore, SERBP1 also co-localizes with TIA1, after arsenite treatment. This migration of SERBP1 to the stress granules is affected by its methylation. Cells pre-treated with Adox, before the arsenite challenge, showed a decrease in SERBP1 recruitment to stress granules, but later also showed high retention of it in the granules[27].
FUNCTIONAL ASPECTS OF HABP4 AND SERBP1
HABP4 and SERBP1: Interaction partners and functions
Frequently, HABP4 and SERBP1 are described as hubs in signaling networks[13], as they can bind to their targets through multiple sites, facilitating the dynamic assembly of complexes. These multiple sites also allow for allosteric responses in biological signaling[43,44]. Besides the ability to have multiple interaction partners, IUPs are targets of PTMs, often having clusters of modifications[45].
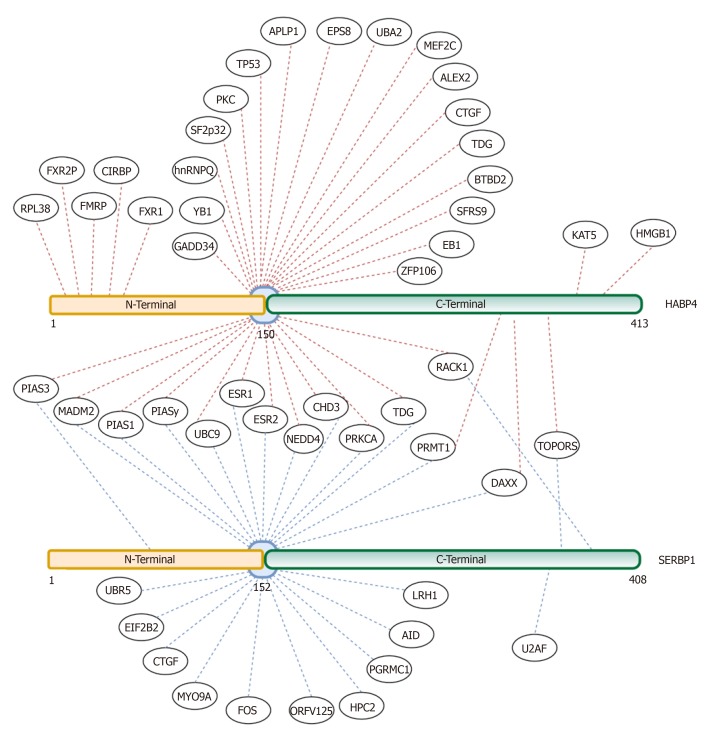
The Y2H system is a powerful tool used to access the pairwise interaction of proteins[5,26,33,34,46]. Such assays with SERBP1 as bait identified several nuclear proteins as interactors and many of them are related to the transcriptional control of gene expression, such as CHD3, DAXX, TOPORS, PIAS1, and PIAS3 (Figure 4)[5,33].
Figure 4.
Global protein interaction network of hyaluronic acid binding protein 4 and serpin mRNA binding protein 1. The published interactors of hyaluronic acid binding protein 4 (HABP4) (top) and serpin mRNA binding protein 1 (SERBP1) (below) were linked to the specific domains of interaction. Proteins that interact with both N- and C-terminal domains, or the site of interaction is unknown, were linked to the blue center. Common interactors between HABP4 and SERBP1 are connected to both proteins and are located between both proteins. Red dotted lines are published interactors of HABP4, and blue dotted lines are interactors of SERBP1. HABP4: Hyaluronic acid binding protein 4; SERBP1: Serpin mRNA binding protein 1.
By using the N- and C- terminal domains of HABP4 as bait, a protein-protein interaction network revealed the pleiotropic functions of this protein. Many of its partners act at different levels of signaling processes (RACK1, PRMT1, EB-1, RIL, ALEX 2, CTGF and EPS8, APLP1, Myosin IXA). Other interactors include DNA binding factors and transcription regulators (such as CHD3, TOPORS, ZFP106, ZFP189, TIP60, BTBD2, YB-1/NSEP1, GADD34, DAXX, PIAS, p100 and HMG). A third category includes RNA metabolism-associated proteins (CIRBP, YB-1 /NSEP1, SFRS9 and SF2 / p32, FXRP). All this evidence seems to suggest that HABP4 has functions in all of the above related biological processes (Figure 4).
Interestingly, at the time of the initial discovery of the Ki-1/57 antigen in 1992[3] a series of cellular localization studies, using electron microscopy, with the original Ki-1 antibody, were performed using mostly the L540 Hodgkin analogous cells. Part of the data were published, but some figures were criticized as potentially nuclear “artifacts” (Figure 5). With today’s knowledge of Ki-1/57/ HABP4 interaction with a number of transcriptional regulators (p53, p100, HMG, Topors, Daxx) and chromatin remodeling machines (CHD3), the images obtained may be interpreted, although in a speculative way, in a different light. Is it possible that the Ki-1 gold-labeling observed represents a chromosome, or chromatin? Is it transcriptionally active or in a repressed state? Future studies should address which parts of the genome are regulated by HABP4.
Figure 5.
Electron microscopic image of a L540 Hodgkin-derived cell labeled with 15 nm gold-particle-coupled Ki-1 antibody (× 29.000 fold magnification). Some labeling on the cell surface is due to Ki-1 binding to the CD30 cell surface receptor. Strong hyaluronic acid binding protein 4 (HABP4) labeling can be detected in the nucleus and the format of the large macrostructure seems to represent part of a chromosome or also (transcriptionally active?) chromatin. A part of the upper end of the macrostructure seems to follow a “corkscrew”-like pattern. Each small black dot is a single Ki-1 gold labeled antibody. Please see[3] for more experimental details. In brief, cells were pelleted by centrifugation. The cell pellet was solidified on ice and the solid cell block was prepared in slices of 2 mm that were dehydrated by incubation in a solution containing stepwise increasing sucrose concentration up to 70%. After fixation in frozen nitrogen, ultrathin cryosections were prepared with an ultracut E, FC-4D and collected on nickel grids. After incubation with Ki-1 antibody, washing, and incubation with secondary reagent Staphylococcal aureus protein A-gold labeled, and further washing, the sections were fixed and contrasted with 4% uranyl acetate to be subsequently analyzed in a Siemens Elmiskop 101, Electron Microscope. We would like to thank Prof. Dr. Hilmar Lemke (Kiel, Germany) for generously providing the electron micrography.
Based on the protein interaction profile and other assays, SERBP1 and HABP4 were functionally related to the regulation of gene expression[5,26,34,36,46]. To identify the candidate genes regulated by SERBP1 and HABP4 over-expression, a global gene expression (DNA microarray, Affymetrix) analysis was performed and revealed that most of the affected genes are related to apoptosis, proliferation, the cell cycle and mRNA metabolism[47]. After over-expression of both SERBP1 or HABP4 around 90% of the genes were down-regulated and these target genes were related to mRNA metabolism and transcription, suggesting that SERBP1/HABP4 may act mainly as a gene expression repressor[47].
The role of SERBP1 in the progesterone response
The plasminogen activator inhibitor type-1 (PAI-1) is the major physiological inhibitor of fibrinolysis and plays important roles in cell adhesion, migration, and invasion[48]. PAI-1 has been related to tumor vascularization and metastasis and some inhibitors are currently being evaluated in cancer therapy[49].
As mentioned before, SERBP1 was previously called PAI-1 mRNA binding protein 1, because it binds to PAI-1 mRNA, and regulates its stability, thereby causing a decrease in overall PAI-1 protein levels in the cell[7]. SERBP1 binds to an AU-rich element located in the 3´-untranslated region of the PAI-1 mRNA. AU-rich elements have been determined as RNA instability promoting sequence motifs. The SERBP1/AU-rich element interaction may be regulated by Sphingosine 1-phosphate (S1P), a sphingolipid metabolite[50].
PAI-1 and SERBP1 mRNA are both over-expressed in ovarian tumors and the expression is higher in more advanced diseases[51]. In fact, the ovarian hormones progesterone (P4) and 17β-estradiol (E2), both regulate SERBP1 mRNA levels, especially in brain regions. These regions are important for the neuroendocrine control of female reproduction[52]. Additionally, the expression of SERBP1 mRNA was also increased in the hypothalamus, which is also important for female reproduction. The expression of SERBP1 mRNA in the brain correlates with the expression of its target mRNA: Progesterone receptor membrane component-1 (PGRMC1) mRNA[52,53]. PGRMC1 mediates the anti-apoptotic action of P4. The over-expression of SERBP1 increased the anti-apoptotic effects of P4 by 10-fold in spontaneously immortalized granulosa cells[54]. However, its effects seem not to be mediated by direct interaction of SERBP1 with the P4 hormone, as this protein possesses only the hyaluronan-binding region (Figure 2). SERBP1 seems to have allosteric effects on PGRMC1 by binding to its C-terminus, which is distant to the putative P4 binding site but could mediate the actions of P4 by serving as a scaffolding protein or co-activator/regulator of PGRMC1[55]. Besides the well-characterized interaction between SERBP1 and PGRMC1, other partners of SERBP1 and HABP4 are also related to P4- and estrogen-related processes, some of which are listed in Table 1.
Table 1.
Table summarizing the interactions of hyaluronic acid binding protein 4 and serpin mRNA binding protein 1 with proteins related to the hormones progesterone and estradiol
| Gene | Protein | Biological process reference | Interaction reference |
| UBR5 | E3 ubiquitin-protein ligase UBR5 | Progesterone receptor signaling pathway[158] | SERBP1[22] |
| EIF2B2 | Translation initiation factor eIF-2B subunit beta | Ovarian follicle development[159] | SERBP1[22] |
| FOS | Proto-oncogene c-Fos | Response to progesterone stimulus[160] | SERBP1[22] |
| PGRMC1 | Membrane-associated progesterone receptor component 1 | Progesterone receptor signaling pathway[161,162] | SERBP1[149] |
| ESR2 | Estrogen receptor beta | Cellular response to estradiol stimulus[163] | SERBP1, HABP4[22] |
| MDM2 | E3 ubiquitin-protein ligase Mdm2 | Cellular response to progesterone stimulus[164] | SERBP1, HABP4[165] |
| NEDD4 | E3 ubiquitin-protein ligase NEDD4 | Progesterone receptor signaling pathway[166] | SERBP1, HABP4[22] |
| ESR1 | Estrogen receptor | Cellular response to estradiol stimulus[163] | SERBP1, HABP4 [22] |
| PKC | Protein Kinase C | Cellular response to estradiol stimulus[167] | HABP4[20] |
| p53 | Cellular tumor antigen p53 | Cellular response to estradiol stimulus[168] | HABP4[21] |
HABP4 and SERBP1 in the context of the DNA damage response
DNA damage is a constant event in cells due to exposure to exogenous (ultraviolet light, ionizing radiation, chemotherapy, radiotherapy) or endogenous agents (reactive oxygen species, oxidation of bases, formation of adducts)[56-60]. Each type of damage is able to activate different cellular responses, including DNA damage repair, changes in the transcriptional response, triggering of apoptosis, senescence, or activation /blockage of the cell cycle checkpoints[61].
Both HABP4 and SERBP1 interacted with proteins related to the DNA damage response. In response to DNA double-strand breaks, SERBP1-depleted cells showed defects in the activation by phosphorylation of CHK1 and RPA2[62]. The effect of SERBP1 in the Homologous Repair pathway is partially explained by the regulation of CtIP (C-terminal binding protein interacting protein) translation in the S phase, once SERBP1 targets CtIP mRNA, thereby controlling its expression levels[62]. CtIP is important for end resection-mediated double-strand break repair via both the Homology Repair (HR) and Non Homology End Joining (NHEJ) pathways[63].
SERBP1 and HABP4 interact further with UBC9, DAXX and PIAS, all of which are related to DNA damage/repair pathways[33,34,46]. HABP4 also interacted in the Y2H system with GADD34, p53, and YB-1[34,46]. The YB-1 protein under normal conditions is mainly located in the cytoplasm, but after genotoxic stress, either by UV-radiation or by treatment with cisplatin or mitomycin C, the protein is translocated to the nucleus, where it has a high affinity for damaged DNA sites[64,65]. In addition, YB-1 interacts with key proteins of the base excision repair (BER) and nucleotide excision repair (NER) pathways, such as APE1, DNA polymerase β and DNA glycosylases[66-68].
The Δ113 isoform of p53, another HABP4 partner, antagonizes the effects of p53 towards apoptotic activity and its expression is increased after irradiation, promoting the repair of the DNA double-strand breaks through the HR and NHEJ pathways. Δ113p53/Δ133p53 promotes repair by regulating the expression of RAD51, LIG4 and RAD52[69]. p53 levels are also regulated by the ubiquitin ligase MDM2, which in turn is regulated by the DAXX protein, that also interacts with HABP4. After treatment with etoposide, ATM phosphorylates DAXX and the complex DAXX-MDM2 is broken down, thereby promoting the activation of p53[70].
Other HABP4 interactors also have functions related to DNA damage. PRMT1, for example, has a function related to the DNA damage response, once it accumulates in cytoplasmic bodies responsive to DNA damage[71]. The protein inhibitor of activated STAT (PIAS) also interacts with HABP4 and when over-expressed in HeLa cells promoted higher resistance to ionizing radiation through its involvement in the repair pathways HR and NHEJ[72]. Additionally, PIAS3 acts as E3 ligase and mediates the required BRCA1 SUMOylation, which in turn is essential for the ubiquitin ligase activity of BRCA1 itself[73].
The emerging regulation of SERBP1 through miRNAs
The microRNAs (miRNAs) are small non-coding RNAs that modulate gene expression by binding to target mRNA[74]. By binding in the 3’ -untranslated region of target mRNAs they inhibit translation or promote degradation of the mRNA. As a consequence the protein expression or non-coding RNAs may be regulated[75]. miRNAs have been explored as biomarkers and therapeutic targets for many pathological conditions including cancer[76], diabetes[77], viral infections[78], cardiovascular disease[79], and neurodegenerative diseases[74], among others.
The study of the regulation of SERPB1 by miRNA has emerged in the past few years. The analysis of miRNA expression in peripheral blood of patients with osteonecrosis of the femoral head revealed that many miRNAs target SERBP1 and p53[80]. In hepatocellular carcinoma (HCC) SERBP1 is regulated by miR-218[81]. miR-218 plays a role as a tumor suppressor in certain types of human cancers and is involved in biological processes such as tumor initiation, progression and metastasis[82]. Despite the negative correlation between the expression of miR-218 and SERBP1 in HCC tissues, when both are co-transfected, miR-218´s ability to inhibit metastasis and reverse Epithelial Mesenchymal Transition, was abolished, suggesting that the function of miR-218 is dependent on SERBP1[81]. In pancreatic cancer cells, miRNAs also act on SERPB1, mainly through miR-448, in a pathway dependent on the long non-coding RNA-PVT1, which regulates proliferation and migration[83].
HABP4 AND SERBP1 INVOLVEMENT IN CANCER
HABP4 and SERBP1 in cancer
Based on the broad clinical spectrum and the underlying, associated molecular behavior, cancer is an extremely complex disease. Tumor initiation and progression result from inherited or acquired genomic alterations within the cells[84], resulting in the acquisition of advantageous features that lead to uncontrolled growth and proliferation[85]. For each type of cancer, specific proteins are frequently altered during cancer initiation and progression, disrupting or promoting protein-protein interactions[86], or causing loss- or gain of function protein variants. The interaction profiles of HABP4 and SERBP1 suggest that they are involved in important cellular events which are related to tumorigenesis[47]. This includes the regulation of gene transcription, translation, RNA splicing, mRNA metabolism, mitotic cell cycle, and apoptosis, as described before[5,22,46,47].
The over-expression of HABP4 in HEK293 cells followed by microarray analysis, showed alterations in gene expression related to proliferation, including those coding for the proteins PLAU, PDXK, NRG1 and α-taxilin (TXLNA). Repression of the TXLNA protein correlates with the proliferative activity of HCC and the metastatic and invasive potential of renal cell carcinoma[87,88]. The over-expression of SERBP1 also resulted in reduced expression of genes related to proliferation, such as GNB1 and CCL14. Based on these findings, both HABP4 and SERBP1 are involved in repressive control mechanisms of proliferation[47]. The proliferation status of cells with HABP4 and SERBP1 over-expression was also evaluated by MTS and EdU incorporation and a lower rate of proliferation was observed, relative to non-transfected cells.
In the same study, HABP4 over-expression led to down-regulation of negative apoptosis regulators, including MAP2K5, ADNP, ANXA4, as well as the positive regulator BCLAF1[47]. In addition, under treatment with the ER-stress inducer thapsigargin, it was found that HABP4 over-expression increased the mRNA level of HSP90B1, an endoplasmic reticulum chaperone that protects against apoptosis[89]. In cells with SERBP1 over-expression, down-regulation of negative apoptotic regulators MAP2K5, PAK7 and FOXO1[47], was observed. The expression of some genes related to the cell cycle or cell division control, such as cyclin-dependent kinase 15 (CDK15), MAPK12[90,91], and the nuclear distribution genes NDEL1 and GORASP1[92,93], was also down-regulated by HABP4 and SERBP1 over-expression, respectively[47].
Taken together, these data suggest that HABP4 and SERBP1 have similar functions, mainly as repressors of genes involved in proliferation, cell cycle and apoptosis regulation, cellular processes that are frequently de-regulated in cancer[47]. However, the target genes can be different for both proteins and consequently the outcome. Several studies suggest important functions of the HABP4 and SERBP1 proteins in human cells that can contribute to a better understanding of the tumorigenic process[47].
HABP4 and SERBP1 are related to epigenetic modifications
Epigenetic modifications, such as alterations in chromatin structure, represent critical regulatory events for cellular proliferation and tumor formation. Many tissue types have altered epigenetic profiles, which contribute to cancer development[94,95]. Chromatin remodeling is one of the epigenetic alterations that is important for the maintenance of chromatin structure and genomic stability[95]. It can change the gene expression patterns and plays important roles in tumor growth, coordinating the transcription factors and protein complexes during the regulation of gene expression[96]. HABP4 and SERBP1 interact with the chromatin remodeling protein “Chromo-Helicase DNA-binding domain-3” (CHD-3), which regulates gene transcription[5,33]. Although the exact mechanisms by which the complexes HABP4- and SERBP1-CHD-3 act, are still unknown, they might influence gene expression regulation of genes related to tumorigenesis. These mechanisms require further analysis. However, the altered transcriptional regulation through pathways targeting chromatin-remodeling has already been reported in tumors[95,97].
The histone modification patterns are crucial for the organization and maintenance of chromatin and as a result, transcriptional regulation. These processes are fundamental in understanding epigenetic mechanisms and their involvement in cancer development, as transcriptional control is essential for appropriate cell proliferation and differentiation[98,99].
Furthermore, HABP4 over-expression in HEK293 cells revealed an increase in the expression in histone genes[47]. These histones could be involved in chromatin compaction during the gene repression process caused by HABP4 over-expression. The above-mentioned repressed genes may also be associated to other, already mentioned, biological processes, such as cellular proliferation, apoptosis and the cell cycle[47,100].
HABP4 may regulate gene expression in response to stress[47]. Some of the histones triggered by HABP4 over-expression are related to the stress response. For example, H2AX, which is responsible for recruiting multiple proteins to chromatin, during the DNA damage/repair response[101]. Additionally, HABP4 and SERBP1 over-expression also influence some histone gene clusters that are preferentially associated with PML-NBs, which respond to basic physiological processes and several forms of stress[102]. Interestingly, PML-NBs also play a role in chromatin regulation and contain histone-modifying enzymes and transcription factors within them[102].
For example, DAXX, a histone component of PML-NB that interacts with HABP4 and SERBP1, possibly coordinates chromatin dynamics by binding to histone deacetylases and chromatin remodeling proteins[103]. Also, HABP4 over-expression reduces histone-lysine N-methyltransferase (SETMAR) expression, which is responsible for the methylation of lysine residues in histone and heterochromatin formation[104].
The histone methyltransferases (HMTs) transfer methyl groups from S-adenosyl methionine (AdoMet) to the lysine and arginine residues of target substrates, which may affect gene transcription, chromatin compaction and effector protein binding[105-107]. HMTs have been found to play fundamental roles in cell differentiation, gene regulation, DNA recombination and DNA damage repair[108-110]. The over-expression of different HMTs and their interaction with oncogenes is associated with the cancer phenotype[111]. Interestingly, HABP4 and SERBP1 not only interact with HMTs but are also methylated by the protein arginine methyltransferase PRMT1[26]. A misregulation of methyltransferases modifies the balance of transcription and leads to changes in cell destination, which in turn may result in tumor development[107]. Further studies are required to better understand the roles of HABP4 and SERBP1 proteins in tumorigenesis events mediated through methyltransferases such as PRMT1 and SETMAR.
HABP4 and cancer
Kobarg and co-workers[112] showed, for the first time, the expression of HABP4 in some tumor cell lines and in activated leukocytes. The HABP4 gene was mapped in the human chromosome 9, bands 9q22.3-31, an area associated with secondary chromosomal aberrations in acute myeloid leukemia and in colon neoplasia[113]. Although previous experiments with HABP4 protein were performed in the Hodgkin’s lymphoma analogous cell line L540[5,112], the relationship between HABP4 and Hodgkin's disease is still not clear[47].
However, in the colon, the 9q22.2-31.2 region, which contains the HABP4 gene, was found to be in linkage disequilibrium with SNP haplotypes found in families with a certain colon neoplasia risk[113]. Preliminary studies have revealed that HABP4 has characteristics also very common in several onco-proteins, such as PTMs, shuttling between the cytoplasm and nucleus, and transcriptional regulation activity[5,40,112,114]. Additionally, HABP4 interacted with PKC[3,21,112], which is considered a tumor suppressor, and loss of function mutations were linked to breast, bladder, skin, and other forms of cancer[115]. However, the exact role of HABP4 in human cells and cancer remains unknown[46].
Another correlation of HABP4 protein with cancer is based on the fact that it belongs to the class of IUPs[8]. Members of the IUPs have received considerable attention lately, due to some of them being involved in the development of several pathologies[46,116]. Indeed, many proteins or domains that are functionally associated with cancer and other human diseases have long disordered regions[116-118]. Additionally, HABP4 plays a role in important regulatory mechanisms such as transcription, translation, cell-cycle checkpoints, and signal transduction, through its interacting proteins and via DNA/RNA either directly or mediated by other proteins[8]. This suggests its functional plasticity and ability to bind to various partners involved in the tumorigenic process[8,116].
The p53 is a nuclear transcription factor that regulates numerous target genes involved in important cellular processes, such as cell cycle arrest and monitoring of the G1 checkpoint, apoptosis, senescence, repair of damaged DNA, as well as metabolic regulation, playing a central role in human cancer as a tumor suppressor[119,120]. When DNA is damaged, free p53 is induced to accumulate in the cell nucleus, mediated through PTMs such as phosphorylation and acetylation[121]. Some studies have shown that the p53 gene is mutated in over 50% of human cancers. The tumor suppressor p53 has also been associated with PML-NBs[32,122], which play significant roles in genome maintenance[37]. Y2H system assays have shown that HABP4 interacts with p53 and with other p53 interacting proteins, as well as with several nuclear proteins involved in the regulation of transcription in the human Hodgkin’s disease analogous cell line L540[32]. HABP4 can negatively influence p53-dependent transcription by blocking its DNA binding. The p53/HABP4 interaction was reported to be inhibited by in vitro phosphorylation of p53[32].
HABP4 and mRNA splicing in cancer
Pre-mRNA splicing is a post-transcriptional process of the eukaryotic gene expression machinery and consists of the removal of introns and the junction of exons in gene transcripts, leading to mature RNAs[123,124]. Deregulation of the splicing process has been discovered to be a critical contributor to the genesis and development of different types of cancers[125,126]. Mutations were found in diverse types of cancers and are linked to alterations in splicing, regulation of specific transcripts and control of spliceosomal activity[123,127].
Several HABP4 interaction partners are involved in gene expression regulation, at the transcriptional level or pre-mRNA splicing[26,32,40,128]. In this way, HABP4 is a potential candidate that could affect cellular fate and function in cancer[127]. HABP4 can also play an important role in tumorigenic events through pre-mRNA splicing alterations[128]. Following this argument, further studies are required to identify the expression of some specific subsets of mRNA that may be regulated by HABP4 in cancer, considering that altered proteins originating from alternative RNA splicing are promising candidates for the diagnosis and even targets for novel therapeutic strategies[123].
It is worth noting that two protein partners were identified for HABP4 that are involved in the regulation of pre-mRNA splicing and cancer. They are serine/arginine-rich splicing factor 9 (SFRS9), a member of the serine/arginine-rich (SR) protein family and heterogeneous nuclear ribonucleoproteins (such as hnRNPQ)[128-130]. In immunoprecipitation and pull-down assays of HeLa cell extracts, both proteins were pulled down by HABP4, suggesting that these splicing proteins interact specifically with HABP4, probably forming functional complexes in vivo[128].
In addition, these proteins have some similar characteristics with HABP4. SFRS9 protein, for example, is a target of arginine methylation, which is required for its localization and trafficking to mammalian cell nuclei[128]. hnRNP proteins act on pre-mRNA splicing through site-specific binding (Arg ⁄Gly-rich clusters) within the target RNA[131], and HABP4 has several RGG-box (Figure 2A) that are important for the interaction with many RNA-binding proteins that mediate splicing decisions[128,132].
Interestingly, SRSF9 has been considered a proto-oncogene by promoting cell proliferation via β-catenin (key effector of the Wnt signaling pathway). β-catenin accumulation in the cytosol and nucleus was found in colon cancer cell lines, and led to increasing colony formation in SRSF9 over-expressing NIH3T3 cells. Furthermore, when these cells were implanted in nude mice they generated tumors of increased sizes compared to control cells, that do not over-express SRSF9[129].
Moreover, elevated levels of SRSF9 expression were found in glioblastoma, colon adenocarcinoma, squamous cell lung carcinoma and malignant melanoma[129], in cancer tissue arrays, when compared to normal tissues. In addition, SRSF9 is implicated in the proliferation of a bladder cancer cell line via an unknown mechanism and SRSF9 over-expression was found in a clinical bladder cancer sample[133].
The hnRNPQ splicing protein has also been reported to be a proto-oncogene[134]. It can promote cell proliferation and translationally regulates cell cycle-related genes in SW480 colon cancer cells, by translation control. In this way, it may increase cell growth ability during tumor formation. Moreover, both its mRNA and protein levels were found to be elevated in colon tumor tissue, possibly, involving transcriptional or post-transcriptional regulation mechanisms.
The contribution of hnRNPs to the control of splicing site selection has been found for apoptotic genes. It was also reported that there was tight control of the balance between the activities of pro- and anti-apoptotic variants produced by apoptotic peptidase activating factor (APAF-1)[135], Bcl-x[136], Fas[137] and caspases[138]. hnRNPs also have a suppressive effect on DNA damage repair[139]. In general, studies have documented that several hnRNPs are involved in human malignancies and metastasis, being promising biomarkers of lung, head and neck, colon, breast, and pancreatic cancers and acute myeloid leukemia[140-142].
SERBP1 and cancer
The over-expression of SERBP1 was reported in epithelial ovarian cancer, in breast-, colon-, prostate- and lung cancer as well as in glioblastoma[51,143,144-146]. In ovarian carcinoma, over-expression of SERBP1 was associated with higher tumor grades (Grade III vs Grades II and I tumors)[143,147]. Indeed, SERBP1 has prognostic value in ovarian cancer and other solid tumors[51]. Although its exact mechanism of action is not well known, SERBP1 was implicated in tumorigenicity and resistance to anti-cancer drugs[51,62].
In human breast cancer, SERBP1 over-expression was classified as a new prognostic marker[144]. A correlation between the expression of SERBP1 and nuclear P4 receptors was observed in malignant breast epithelial cells[27,144,148]. This relation is very important, as P4 receptors are ligand-activated transcription factors, playing a crucial role in the regulation of growth, survival, and differentiation of normal and malignant breast epithelial cells[144,148]. Furthermore, as pointed out above, SERBP1 interacts with PGRMC1, which is involved in mediating anti-apoptotic actions through the P4 receptor[55,144,149].
Interestingly, abundant SERBP1 expression in human breast cancer was associated with low PAI-1 protein levels in Western blot analysis[144], showing that SERBP1 can not only stabilize but can also destabilize PAI-1 mRNA, depending on the cellular context[7]. Heaton and co-workers documented an inverse relationship between SERBP1 and PAI in rat hepatoma, reporting that high SERBP1 protein levels, lead to increased degradation of PAI-1 mRNA and consecutively to low PAI-1 protein levels in rat hepatoma[150].
Other studies showed over-expression and high protein levels of PAI-1 in breast cancer, which was associated with poor prognosis. PAI-1 is considered a valuable factor in clinical practice[51,144,151]. In ovarian cancer, the over-expression of PAI-1 was also detected and related to advanced tumor stages, and poor prognosis in ovarian cancer patients[51,152-154]. Moreover, studies have reported an association between the high expression of PAI-1 in ovarian cancer and its histological grade[153,155], tumor stage[156], tumor recurrence[153] and residual tumor[157] (Table 2).
Table 2.
Hyaluronic acid binding protein 4 and serpin mRNA binding protein 1 expression in different types of cancer
| Protein | Expression/overexpression | Cancer types | Ref. |
| HABP4 | Expression | Hodgkin’s lymphoma | Kobarg et al[112] |
| HABP4 | Expression | B-cell lymphatic leukemia | Kobarg et al[112] |
| HABP4 | Expression | non-Hodgkin-T-cell lymphoma | Kobarg et al[112] |
| HABP4 | Expression | Bladder | Kobarg et al[112] |
| SERBP1 | Overexpression | Ovarian | Koensgen et al[51] |
| SERBP1 | Overexpression | Breast | Serce et al[144] |
| SERBP1 | Overexpression | Colon | Wang et al[81] |
| SERBP1 | Overexpression | Prostate | Guo et al[145] |
| SERBP1 | Overexpression | Lung | Sun et al[147] |
| SERBP1 | Overexpression | Glioblastoma | Hlavaty et al[146] |
CONCLUSION
HABP4 and SERBP1 share high levels of amino acid sequence identity and similarity and seem to have overlapping functions in the cell, related to transcription regulation, mRNA metabolism and DNA damage and stress responses. However, they have also exclusive interacting partners and might be differentially regulated. Thus, HABP4 and SERBP1 may be required in different situations to exert unique functions specific to each paralog protein. In this review, we presented the emerging role of HABP4 and SERBP1 in the cancer field, and the need for further studies to understand more deeply the cellular functions of both proteins.
Footnotes
Conflict-of-interest statement: The authors have no conflicts of interest to declare.
Manuscript source: Unsolicited manuscript
Peer-review started: July 16, 2019
First decision: August 20, 2019
Article in press: October 15, 2019
Specialty type: Biochemistry and molecular biology
Country of origin: Brazil
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Chen CJ, Tanabe S, Zhou J S-Editor: Ma RY L-Editor: Webster JR E-Editor: Qi LL
Contributor Information
Carolina Colleti, Faculty of Pharmaceutical Sciences, University of Campinas, Campinas 13083-871, Brazil; Institute of Biology, Departament of Biochemistry and Tissue Biology, University of Campinas, Campinas 13083-862, Brazil.
Talita Diniz Melo-Hanchuk, Institute of Biology, Departament of Biochemistry and Tissue Biology, University of Campinas, Campinas 13083-862, Brazil.
Flávia Regina Moraes da Silva, Faculty of Pharmaceutical Sciences, University of Campinas, Campinas 13083-871, Brazil; Institute of Biology, Departament of Biochemistry and Tissue Biology, University of Campinas, Campinas 13083-862, Brazil.
Ângela Saito, Laboratório Nacional de Biociências, CNPEM, Campinas 13083-970, Brazil.
Jörg Kobarg, Faculty of Pharmaceutical Sciences, University of Campinas, Campinas 13083-871, Brazil; Institute of Biology, Departament of Biochemistry and Tissue Biology, University of Campinas, Campinas 13083-862, Brazil. jorgkoba@unicamp.br.
References
- 1.Schwab U, Stein H, Gerdes J, Lemke H, Kirchner H, Schaadt M, Diehl V. Production of a monoclonal antibody specific for Hodgkin and Sternberg-Reed cells of Hodgkin's disease and a subset of normal lymphoid cells. Nature. 1982;299:65–67. doi: 10.1038/299065a0. [DOI] [PubMed] [Google Scholar]
- 2.Hansen H, Lemke H, Bredfeldt G, Könnecke I, Havsteen B. The Hodgkin-associated Ki-1 antigen exists in an intracellular and a membrane-bound form. Biol Chem Hoppe Seyler. 1989;370:409–416. doi: 10.1515/bchm3.1989.370.1.409. [DOI] [PubMed] [Google Scholar]
- 3.Rohde D, Hansen H, Hafner M, Lange H, Mielke V, Hansmann ML, Lemke H. Cellular localizations and processing of the two molecular forms of the Hodgkin-associated Ki-1 (CD30) antigen. The protein kinase Ki-1/57 occurs in the nucleus. Am J Pathol. 1992;140:473–482. [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L, Grammatikakis N, Yoneda M, Banerjee SD, Toole BP. Molecular characterization of a novel intracellular hyaluronan-binding protein. J Biol Chem. 2000;275:29829–29839. doi: 10.1074/jbc.M002737200. [DOI] [PubMed] [Google Scholar]
- 5.Lemos TA, Passos DO, Nery FC, Kobarg J. Characterization of a new family of proteins that interact with the C-terminal region of the chromatin-remodeling factor CHD-3. FEBS Lett. 2003;533:14–20. doi: 10.1016/s0014-5793(02)03737-7. [DOI] [PubMed] [Google Scholar]
- 6.Lai CH, Chou CY, Ch'ang LY, Liu CS, Lin W. Identification of novel human genes evolutionarily conserved in Caenorhabditis elegans by comparative proteomics. Genome Res. 2000;10:703–713. doi: 10.1101/gr.10.5.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Heaton JH, Dlakic WM, Dlakic M, Gelehrter TD. Identification and cDNA cloning of a novel RNA-binding protein that interacts with the cyclic nucleotide-responsive sequence in the Type-1 plasminogen activator inhibitor mRNA. J Biol Chem. 2001;276:3341–3347. doi: 10.1074/jbc.M006538200. [DOI] [PubMed] [Google Scholar]
- 8.Bressan GC, Silva JC, Borges JC, Dos Passos DO, Ramos CH, Torriani IL, Kobarg J. Human regulatory protein Ki-1/57 has characteristics of an intrinsically unstructured protein. J Proteome Res. 2008;7:4465–4474. doi: 10.1021/pr8005342. [DOI] [PubMed] [Google Scholar]
- 9.Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 10.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 11.Jaravine VA, Alexandrescu AT, Grzesiek S. Observation of the closing of individual hydrogen bonds during TFE-induced helix formation in a peptide. Protein Sci. 2001;10:943–950. doi: 10.1110/ps.48501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lawrence JR, Johnson WC. Lifson-Roig nucleation for alpha-helices in trifluoroethanol: context has a strong effect on the helical propensity of amino acids. Biophys Chem. 2002;101-102:375–385. doi: 10.1016/s0301-4622(02)00173-4. [DOI] [PubMed] [Google Scholar]
- 13.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 14.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 15.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 16.Duan G, Walther D. The roles of post-translational modifications in the context of protein interaction networks. PLoS Comput Biol. 2015;11:e1004049. doi: 10.1371/journal.pcbi.1004049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh V, Ram M, Kumar R, Prasad R, Roy BK, Singh KK. Phosphorylation: Implications in Cancer. Protein J. 2017;36:1–6. doi: 10.1007/s10930-017-9696-z. [DOI] [PubMed] [Google Scholar]
- 18.Peng C, Wong CC. The story of protein arginine methylation: characterization, regulation, and function. Expert Rev Proteomics. 2017;14:157–170. doi: 10.1080/14789450.2017.1275573. [DOI] [PubMed] [Google Scholar]
- 19.Ohtake F, Tsuchiya H. The emerging complexity of ubiquitin architecture. J Biochem. 2017;161:125–133. doi: 10.1093/jb/mvw088. [DOI] [PubMed] [Google Scholar]
- 20.Nery FC, Passos DO, Garcia VS, Kobarg J. Ki-1/57 interacts with RACK1 and is a substrate for the phosphorylation by phorbol 12-myristate 13-acetate-activated protein kinase C. J Biol Chem. 2004;279:11444–11455. doi: 10.1074/jbc.M306672200. [DOI] [PubMed] [Google Scholar]
- 21.Nery FC, Bressan GC, Alborghetti MR, Passos DO, Kuniyoshi TM, Ramos CH, Oyama S, Jr, Kobarg J. A spectroscopic analysis of the interaction between the human regulatory proteins RACK1 and Ki-1/57. Biol Chem. 2006;387:577–582. doi: 10.1515/BC.2006.074. [DOI] [PubMed] [Google Scholar]
- 22.Saito Â, Souza EE, Costa FC, Meirelles GV, Gonçalves KA, Santos MT, Bressan GC, McComb ME, Costello CE, Whelan SA, Kobarg J. Human Regulatory Protein Ki-1/57 Is a Target of SUMOylation and Affects PML Nuclear Body Formation. J Proteome Res. 2017;16:3147–3157. doi: 10.1021/acs.jproteome.7b00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hansen H, Bredfeldt G, Havsteen B, Lemke H. Protein kinase activity of the intracellular but not of the membrane-associated form of the Ki-1 antigen (CD30) Res Immunol. 1990;141:13–31. doi: 10.1016/0923-2494(90)90098-j. [DOI] [PubMed] [Google Scholar]
- 24.Bolger GB. The RNA-binding protein SERBP1 interacts selectively with the signaling protein RACK1. Cell Signal. 2017;35:256–263. doi: 10.1016/j.cellsig.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma G, Pallesen J, Das S, Grassucci R, Langlois R, Hampton CM, Kelly DF, des Georges A, Frank J. Affinity grid-based cryo-EM of PKC binding to RACK1 on the ribosome. J Struct Biol. 2013;181:190–194. doi: 10.1016/j.jsb.2012.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passos DO, Bressan GC, Nery FC, Kobarg J. Ki-1/57 interacts with PRMT1 and is a substrate for arginine methylation. FEBS J. 2006;273:3946–3961. doi: 10.1111/j.1742-4658.2006.05399.x. [DOI] [PubMed] [Google Scholar]
- 27.Lee YJ, Wei HM, Chen LY, Li C. Localization of SERBP1 in stress granules and nucleoli. FEBS J. 2014;281:352–364. doi: 10.1111/febs.12606. [DOI] [PubMed] [Google Scholar]
- 28.Lee YJ, Hsieh WY, Chen LY, Li C. Protein arginine methylation of SERBP1 by protein arginine methyltransferase 1 affects cytoplasmic/nuclear distribution. J Cell Biochem. 2012;113:2721–2728. doi: 10.1002/jcb.24151. [DOI] [PubMed] [Google Scholar]
- 29.Srinivasan S, Shankar SR, Wang Y, Taneja R. SUMOylation of G9a regulates its function as an activator of myoblast proliferation. Cell Death Dis. 2019;10:250. doi: 10.1038/s41419-019-1465-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rosonina E, Akhter A, Dou Y, Babu J, Sri Theivakadadcham VS. Regulation of transcription factors by sumoylation. Transcription. 2017;8:220–231. doi: 10.1080/21541264.2017.1311829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richard P, Vethantham V, Manley JL. Roles of Sumoylation in mRNA Processing and Metabolism. Adv Exp Med Biol. 2017;963:15–33. doi: 10.1007/978-3-319-50044-7_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nery FC, Rui E, Kuniyoshi TM, Kobarg J. Evidence for the interaction of the regulatory protein Ki-1/57 with p53 and its interacting proteins. Biochem Biophys Res Commun. 2006;341:847–855. doi: 10.1016/j.bbrc.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 33.Lemos TA, Kobarg J. CGI-55 interacts with nuclear proteins and co-localizes to p80-coilin positive-coiled bodies in the nucleus. Cell Biochem Biophys. 2006;44:463–474. doi: 10.1385/CBB:44:3:463. [DOI] [PubMed] [Google Scholar]
- 34.Bressan GC, Quaresma AJ, Moraes EC, Manfiolli AO, Passos DO, Gomes MD, Kobarg J. Functional association of human Ki-1/57 with pre-mRNA splicing events. FEBS J. 2009;276:3770–3783. doi: 10.1111/j.1742-4658.2009.07092.x. [DOI] [PubMed] [Google Scholar]
- 35.Morimoto M, Boerkoel CF. The role of nuclear bodies in gene expression and disease. Biology (Basel) 2013;2:976–1033. doi: 10.3390/biology2030976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staněk D. Cajal bodies and snRNPs - friends with benefits. RNA Biol. 2017;14:671–679. doi: 10.1080/15476286.2016.1231359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang HR, Munkhjargal A, Kim MJ, Park SY, Jung E, Ryu JH, Yang Y, Lim JS, Kim Y. The functional roles of PML nuclear bodies in genome maintenance. Mutat Res. 2018;809:99–107. doi: 10.1016/j.mrfmmm.2017.05.002. [DOI] [PubMed] [Google Scholar]
- 38.Zhong S, Müller S, Ronchetti S, Freemont PS, Dejean A, Pandolfi PP. Role of SUMO-1-modified PML in nuclear body formation. Blood. 2000;95:2748–2752. [PubMed] [Google Scholar]
- 39.Seker H, Rubbi C, Linke SP, Bowman ED, Garfield S, Hansen L, Borden KL, Milner J, Harris CC. UV-C-induced DNA damage leads to p53-dependent nuclear trafficking of PML. Oncogene. 2003;22:1620–1628. doi: 10.1038/sj.onc.1206140. [DOI] [PubMed] [Google Scholar]
- 40.Gonçalves Kde A, Bressan GC, Saito A, Morello LG, Zanchin NI, Kobarg J. Evidence for the association of the human regulatory protein Ki-1/57 with the translational machinery. FEBS Lett. 2011;585:2556–2560. doi: 10.1016/j.febslet.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 41.Darnell JC, Fraser CE, Mostovetsky O, Darnell RB. Discrimination of common and unique RNA-binding activities among Fragile X mental retardation protein paralogs. Hum Mol Genet. 2009;18:3164–3177. doi: 10.1093/hmg/ddp255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kedersha NL, Gupta M, Li W, Miller I, Anderson P. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 alpha to the assembly of mammalian stress granules. J Cell Biol. 1999;147:1431–1442. doi: 10.1083/jcb.147.7.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Flock T, Weatheritt RJ, Latysheva NS, Babu MM. Controlling entropy to tune the functions of intrinsically disordered regions. Curr Opin Struct Biol. 2014;26:62–72. doi: 10.1016/j.sbi.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 45.Pejaver V, Hsu WL, Xin F, Dunker AK, Uversky VN, Radivojac P. The structural and functional signatures of proteins that undergo multiple events of post-translational modification. Protein Sci. 2014;23:1077–1093. doi: 10.1002/pro.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bressan GC, Kobarg J. From protein interaction profile to functional assignment: the human protein Ki-1/57 is associated with pre-mRNA splicing events. RNA Biol. 2010;7:268–271. doi: 10.4161/rna.7.3.11489. [DOI] [PubMed] [Google Scholar]
- 47.Costa FC, Saito A, Gonçalves KA, Vidigal PM, Meirelles GV, Bressan GC, Kobarg J. Ki-1/57 and CGI-55 ectopic expression impact cellular pathways involved in proliferation and stress response regulation. Biochim Biophys Acta. 2014;1843:2944–2956. doi: 10.1016/j.bbamcr.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 48.Yasar Yildiz S, Kuru P, Toksoy Oner E, Agirbasli M. Functional stability of plasminogen activator inhibitor-1. ScientificWorldJournal. 2014;2014:858293. doi: 10.1155/2014/858293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li S, Wei X, He J, Tian X, Yuan S, Sun L. Plasminogen activator inhibitor-1 in cancer research. Biomed Pharmacother. 2018;105:83–94. doi: 10.1016/j.biopha.2018.05.119. [DOI] [PubMed] [Google Scholar]
- 50.Iwaki S, Yamamura S, Asai M, Sobel BE, Fujii S. Posttranscriptional regulation of expression of plasminogen activator inhibitor type-1 by sphingosine 1-phosphate in HepG2 liver cells. Biochim Biophys Acta. 2012;1819:1132–1141. doi: 10.1016/j.bbagrm.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 51.Koensgen D, Mustea A, Klaman I, Sun P, Zafrakas M, Lichtenegger W, Denkert C, Dahl E, Sehouli J. Expression analysis and RNA localization of PAI-RBP1 (SERBP1) in epithelial ovarian cancer: association with tumor progression. Gynecol Oncol. 2007;107:266–273. doi: 10.1016/j.ygyno.2007.06.023. [DOI] [PubMed] [Google Scholar]
- 52.Intlekofer KA, Petersen SL. Distribution of mRNAs encoding classical progestin receptor, progesterone membrane components 1 and 2, serpine mRNA binding protein 1, and progestin and ADIPOQ receptor family members 7 and 8 in rat forebrain. Neuroscience. 2011;172:55–65. doi: 10.1016/j.neuroscience.2010.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Intlekofer KA, Petersen SL. 17β-estradiol and progesterone regulate multiple progestin signaling molecules in the anteroventral periventricular nucleus, ventromedial nucleus and sexually dimorphic nucleus of the preoptic area in female rats. Neuroscience. 2011;176:86–92. doi: 10.1016/j.neuroscience.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Peluso JJ, Pappalardo A, Fernandez G, Wu CA. Involvement of an unnamed protein, RDA288, in the mechanism through which progesterone mediates its antiapoptotic action in spontaneously immortalized granulosa cells. Endocrinology. 2004;145:3014–3022. doi: 10.1210/en.2004-0067. [DOI] [PubMed] [Google Scholar]
- 55.Peluso JJ, Romak J, Liu X. Progesterone receptor membrane component-1 (PGRMC1) is the mediator of progesterone's antiapoptotic action in spontaneously immortalized granulosa cells as revealed by PGRMC1 small interfering ribonucleic acid treatment and functional analysis of PGRMC1 mutations. Endocrinology. 2008;149:534–543. doi: 10.1210/en.2007-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boulton S, Kyle S, Durkacz BW. Mechanisms of enhancement of cytotoxicity in etoposide and ionising radiation-treated cells by the protein kinase inhibitor wortmannin. Eur J Cancer. 2000;36:535–541. doi: 10.1016/s0959-8049(99)00311-1. [DOI] [PubMed] [Google Scholar]
- 57.Henle ES, Linn S. Formation, prevention, and repair of DNA damage by iron/hydrogen peroxide. J Biol Chem. 1997;272:19095–19098. doi: 10.1074/jbc.272.31.19095. [DOI] [PubMed] [Google Scholar]
- 58.Pommier Y, Redon C, Rao VA, Seiler JA, Sordet O, Takemura H, Antony S, Meng L, Liao Z, Kohlhagen G, Zhang H, Kohn KW. Repair of and checkpoint response to topoisomerase I-mediated DNA damage. Mutat Res. 2003;532:173–203. doi: 10.1016/j.mrfmmm.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 59.Todd RC, Lippard SJ. Inhibition of transcription by platinum antitumor compounds. Metallomics. 2009;1:280–291. doi: 10.1039/b907567d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature. 2000;408:433–439. doi: 10.1038/35044005. [DOI] [PubMed] [Google Scholar]
- 61.Feringa FM, Raaijmakers JA, Hadders MA, Vaarting C, Macurek L, Heitink L, Krenning L, Medema RH. Persistent repair intermediates induce senescence. Nat Commun. 2018;9:3923. doi: 10.1038/s41467-018-06308-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahn JW, Kim S, Na W, Baek SJ, Kim JH, Min K, Yeom J, Kwak H, Jeong S, Lee C, Kim SY, Choi CY. SERBP1 affects homologous recombination-mediated DNA repair by regulation of CtIP translation during S phase. Nucleic Acids Res. 2015;43:6321–6333. doi: 10.1093/nar/gkv592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang H, Qiu Z, Liu B, Wu Y, Ren J, Liu Y, Zhao Y, Wang Y, Hao S, Li Z, Peng B, Xu X. PLK1 targets CtIP to promote microhomology-mediated end joining. Nucleic Acids Res. 2018;46:10724–10739. doi: 10.1093/nar/gky810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cohen SB, Ma W, Valova VA, Algie M, Harfoot R, Woolley AG, Robinson PJ, Braithwaite AW. Genotoxic stress-induced nuclear localization of oncoprotein YB-1 in the absence of proteolytic processing. Oncogene. 2010;29:403–410. doi: 10.1038/onc.2009.321. [DOI] [PubMed] [Google Scholar]
- 65.Koike K, Uchiumi T, Ohga T, Toh S, Wada M, Kohno K, Kuwano M. Nuclear translocation of the Y-box binding protein by ultraviolet irradiation. FEBS Lett. 1997;417:390–394. doi: 10.1016/s0014-5793(97)01296-9. [DOI] [PubMed] [Google Scholar]
- 66.Chattopadhyay R, Das S, Maiti AK, Boldogh I, Xie J, Hazra TK, Kohno K, Mitra S, Bhakat KK. Regulatory role of human AP-endonuclease (APE1/Ref-1) in YB-1-mediated activation of the multidrug resistance gene MDR1. Mol Cell Biol. 2008;28:7066–7080. doi: 10.1128/MCB.00244-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim HL, Koedrith P, Lee SM, Kim YJ, Seo YR. Base excision DNA repair defect in thioredoxin-1 (Trx1)-deficient cells. Mutat Res. 2013;751-752:1–7. doi: 10.1016/j.mrfmmm.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 68.Prasad R, Williams JG, Hou EW, Wilson SH. Pol β associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 2012;40:11571–11582. doi: 10.1093/nar/gks898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gong L, Gong H, Pan X, Chang C, Ou Z, Ye S, Yin L, Yang L, Tao T, Zhang Z, Liu C, Lane DP, Peng J, Chen J. p53 isoform Δ113p53/Δ133p53 promotes DNA double-strand break repair to protect cell from death and senescence in response to DNA damage. Cell Res. 2015;25:351–369. doi: 10.1038/cr.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang J, Agrawal T, Cheng Q, Qu L, Brewer MD, Chen J, Yang X. Phosphorylation of Daxx by ATM contributes to DNA damage-induced p53 activation. PLoS One. 2013;8:e55813. doi: 10.1371/journal.pone.0055813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suchánková J, Legartová S, Sehnalová P, Kozubek S, Valente S, Labella D, Mai A, Eckerich C, Fackelmayer FO, Sorokin DV, Bartova E. PRMT1 arginine methyltransferase accumulates in cytoplasmic bodies that respond to selective inhibition and DNA damage. Eur J Histochem. 2014;58:2389. doi: 10.4081/ejh.2014.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu S, Fan Z, Geng Z, Zhang H, Ye Q, Jiao S, Xu X. PIAS3 promotes homology-directed repair and distal non-homologous end joining. Oncol Lett. 2013;6:1045–1048. doi: 10.3892/ol.2013.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Morris JR, Boutell C, Keppler M, Densham R, Weekes D, Alamshah A, Butler L, Galanty Y, Pangon L, Kiuchi T, Ng T, Solomon E. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature. 2009;462:886–890. doi: 10.1038/nature08593. [DOI] [PubMed] [Google Scholar]
- 74.Joilin G, Leigh PN, Newbury SF, Hafezparast M. An Overview of MicroRNAs as Biomarkers of ALS. Front Neurol. 2019;10:186. doi: 10.3389/fneur.2019.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vislovukh A, Vargas TR, Polesskaya A, Groisman I. Role of 3'-untranslated region translational control in cancer development, diagnostics and treatment. World J Biol Chem. 2014;5:40–57. doi: 10.4331/wjbc.v5.i1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pardini B, Calin GA. MicroRNAs and Long Non-Coding RNAs and Their Hormone-Like Activities in Cancer. Cancers (Basel) 2019;11 doi: 10.3390/cancers11030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kadamkode V, Banerjee G. Micro RNA: an epigenetic regulator of type 2 diabetes. Microrna. 2014;3:86–97. doi: 10.2174/2211536603666141118232514. [DOI] [PubMed] [Google Scholar]
- 78.Lv J, Zhang Z, Pan L, Zhang Y. MicroRNA-34/449 family and viral infections. Virus Res. 2019;260:1–6. doi: 10.1016/j.virusres.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Moghaddam AS, Afshari JT, Esmaeili SA, Saburi E, Joneidi Z, Momtazi-Borojeni AA. Cardioprotective microRNAs: Lessons from stem cell-derived exosomal microRNAs to treat cardiovascular disease. Atherosclerosis. 2019;285:1–9. doi: 10.1016/j.atherosclerosis.2019.03.016. [DOI] [PubMed] [Google Scholar]
- 80.Kao GS, Tu YK, Sung PH, Wang FS, Lu YD, Wu CT, Lin RLC, Yip HK, Lee MS. MicroRNA-mediated interacting circuits predict hypoxia and inhibited osteogenesis of stem cells, and dysregulated angiogenesis are involved in osteonecrosis of the femoral head. Int Orthop. 2018;42:1605–1614. doi: 10.1007/s00264-018-3895-x. [DOI] [PubMed] [Google Scholar]
- 81.Wang T, Xu L, Jia R, Wei J. MiR-218 suppresses the metastasis and EMT of HCC cells via targeting SERBP1. Acta Biochim Biophys Sin (Shanghai) 2017;49:383–391. doi: 10.1093/abbs/gmx017. [DOI] [PubMed] [Google Scholar]
- 82.Lu YF, Zhang L, Waye MM, Fu WM, Zhang JF. MiR-218 mediates tumorigenesis and metastasis: Perspectives and implications. Exp Cell Res. 2015;334:173–182. doi: 10.1016/j.yexcr.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 83.Zhao L, Kong H, Sun H, Chen Z, Chen B, Zhou M. LncRNA-PVT1 promotes pancreatic cancer cells proliferation and migration through acting as a molecular sponge to regulate miR-448. J Cell Physiol. 2018;233:4044–4055. doi: 10.1002/jcp.26072. [DOI] [PubMed] [Google Scholar]
- 84.Chik F, Szyf M, Rabbani SA. Role of epigenetics in cancer initiation and progression. Adv Exp Med Biol. 2011;720:91–104. doi: 10.1007/978-1-4614-0254-1_8. [DOI] [PubMed] [Google Scholar]
- 85.Imran A, Qamar HY, Ali Q, Naeem H, Riaz M, Amin S, Kanwal N, Ali F, Sabar MF, Nasir IA. Role of Molecular Biology in Cancer Treatment: A Review Article. Iran J Public Health. 2017;46:1475–1485. [PMC free article] [PubMed] [Google Scholar]
- 86.Climente-González H, Porta-Pardo E, Godzik A, Eyras E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017;20:2215–2226. doi: 10.1016/j.celrep.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 87.Ohtomo N, Tomiya T, Tanoue Y, Inoue Y, Nishikawa T, Ikeda H, Seyama Y, Kokudo N, Shibahara J, Fukayama M, Koike K, Shirataki H, Fujiwara K. Expression of α-taxilin in hepatocellular carcinoma correlates with growth activity and malignant potential of the tumor. Int J Oncol. 2010;37:1417–1423. doi: 10.3892/ijo_00000793. [DOI] [PubMed] [Google Scholar]
- 88.Mashidori T, Shirataki H, Kamai T, Nakamura F, Yoshida K. Increased alpha-taxilin protein expression is associated with the metastatic and invasive potential of renal cell cancer. Biomed Res. 2011;32:103–110. doi: 10.2220/biomedres.32.103. [DOI] [PubMed] [Google Scholar]
- 89.Pan Z, Erkan M, Streit S, Friess H, Kleeff J. Silencing of GRP94 expression promotes apoptosis in pancreatic cancer cells. Int J Oncol. 2009;35:823–828. doi: 10.3892/ijo_00000395. [DOI] [PubMed] [Google Scholar]
- 90.Zhao F, Vilardi A, Neely RJ, Choi JK. Promotion of cell cycle progression by basic helix-loop-helix E2A. Mol Cell Biol. 2001;21:6346–6357. doi: 10.1128/MCB.21.18.6346-6357.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wang X, McGowan CH, Zhao M, He L, Downey JS, Fearns C, Wang Y, Huang S, Han J. Involvement of the MKK6-p38gamma cascade in gamma-radiation-induced cell cycle arrest. Mol Cell Biol. 2000;20:4543–4552. doi: 10.1128/mcb.20.13.4543-4552.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liang Y, Yu W, Li Y, Yu L, Zhang Q, Wang F, Yang Z, Du J, Huang Q, Yao X, Zhu X. Nudel modulates kinetochore association and function of cytoplasmic dynein in M phase. Mol Biol Cell. 2007;18:2656–2666. doi: 10.1091/mbc.E06-04-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sütterlin C, Polishchuk R, Pecot M, Malhotra V. The Golgi-associated protein GRASP65 regulates spindle dynamics and is essential for cell division. Mol Biol Cell. 2005;16:3211–3222. doi: 10.1091/mbc.E04-12-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ando M, Saito Y, Xu G, Bui NQ, Medetgul-Ernar K, Pu M, Fisch K, Ren S, Sakai A, Fukusumi T, Liu C, Haft S, Pang J, Mark A, Gaykalova DA, Guo T, Favorov AV, Yegnasubramanian S, Fertig EJ, Ha P, Tamayo P, Yamasoba T, Ideker T, Messer K, Califano JA. Chromatin dysregulation and DNA methylation at transcription start sites associated with transcriptional repression in cancers. Nat Commun. 2019;10:2188. doi: 10.1038/s41467-019-09937-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaur J, Daoud A, Eblen ST. Targeting Chromatin Remodeling for Cancer Therapy. Curr Mol Pharmacol. 2019;12:215–229. doi: 10.2174/1874467212666190215112915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sadikovic B, Al-Romaih K, Squire JA, Zielenska M. Cause and consequences of genetic and epigenetic alterations in human cancer. Curr Genomics. 2008;9:394–408. doi: 10.2174/138920208785699580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shankar S, Kumar D, Srivastava RK. Epigenetic modifications by dietary phytochemicals: implications for personalized nutrition. Pharmacol Ther. 2013;138:1–17. doi: 10.1016/j.pharmthera.2012.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Patra S, Panigrahi DP, Praharaj PP, Bhol CS, Mahapatra KK, Mishra SR, Behera BP, Jena M, Bhutia SK. Dysregulation of histone deacetylases in carcinogenesis and tumor progression: a possible link to apoptosis and autophagy. Cell Mol Life Sci. 2019;76:3263–3282. doi: 10.1007/s00018-019-03098-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mohammad HP, Barbash O, Creasy CL. Targeting epigenetic modifications in cancer therapy: erasing the roadmap to cancer. Nat Med. 2019;25:403–418. doi: 10.1038/s41591-019-0376-8. [DOI] [PubMed] [Google Scholar]
- 100.Hake SB, Garcia BA, Duncan EM, Kauer M, Dellaire G, Shabanowitz J, Bazett-Jones DP, Allis CD, Hunt DF. Expression patterns and post-translational modifications associated with mammalian histone H3 variants. J Biol Chem. 2006;281:559–568. doi: 10.1074/jbc.M509266200. [DOI] [PubMed] [Google Scholar]
- 101.Lee YY, Yu YB, Gunawardena HP, Xie L, Chen X. BCLAF1 is a radiation-induced H2AX-interacting partner involved in γH2AX-mediated regulation of apoptosis and DNA repair. Cell Death Dis. 2012;3:e359. doi: 10.1038/cddis.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Salomoni P. The PML-Interacting Protein DAXX: Histone Loading Gets into the Picture. Front Oncol. 2013;3:152. doi: 10.3389/fonc.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sudharsan R, Azuma Y. The SUMO ligase PIAS1 regulates UV-induced apoptosis by recruiting Daxx to SUMOylated foci. J Cell Sci. 2012;125:5819–5829. doi: 10.1242/jcs.110825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cho S, Park JS, Kang YK. Dual functions of histone-lysine N-methyltransferase Setdb1 protein at promyelocytic leukemia-nuclear body (PML-NB): maintaining PML-NB structure and regulating the expression of its associated genes. J Biol Chem. 2011;286:41115–41124. doi: 10.1074/jbc.M111.248534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Collazo E, Couture JF, Bulfer S, Trievel RC. A coupled fluorescent assay for histone methyltransferases. Anal Biochem. 2005;342:86–92. doi: 10.1016/j.ab.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 106.Chaib H, Prébet T, Vey N, Collette Y. [Histone methyltransferases: a new class of therapeutic targets in cancer treatment?] Med Sci (Paris) 2011;27:725–732. doi: 10.1051/medsci/2011278014. [DOI] [PubMed] [Google Scholar]
- 107.Li J, Zhu S, Ke XX, Cui H. Role of several histone lysine methyltransferases in tumor development. Biomed Rep. 2016;4:293–299. doi: 10.3892/br.2016.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 110.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. Nat Rev Immunol. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 111.Deb M, Kar S, Sengupta D, Shilpi A, Parbin S, Rath SK, Londhe VA, Patra SK. Chromatin dynamics: H3K4 methylation and H3 variant replacement during development and in cancer. Cell Mol Life Sci. 2014;71:3439–3463. doi: 10.1007/s00018-014-1605-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kobarg J, Schnittger S, Fonatsch C, Lemke H, Bowen MA, Buck F, Hansen HP. Characterization, mapping and partial cDNA sequence of the 57-kD intracellular Ki-1 antigen. Exp Clin Immunogenet. 1997;14:273–280. [PubMed] [Google Scholar]
- 113.Gray-McGuire C, Guda K, Adrianto I, Lin CP, Natale L, Potter JD, Newcomb P, Poole EM, Ulrich CM, Lindor N, Goode EL, Fridley BL, Jenkins R, Le Marchand L, Casey G, Haile R, Hopper J, Jenkins M, Young J, Buchanan D, Gallinger S, Adams M, Lewis S, Willis J, Elston R, Markowitz SD, Wiesner GL. Confirmation of linkage to and localization of familial colon cancer risk haplotype on chromosome 9q22. Cancer Res. 2010;70:5409–5418. doi: 10.1158/0008-5472.CAN-10-0188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kobarg CB, Kobarg J, Crosara-Alberto DP, Theizen TH, Franchini KG. MEF2C DNA-binding activity is inhibited through its interaction with the regulatory protein Ki-1/57. FEBS Lett. 2005;579:2615–2622. doi: 10.1016/j.febslet.2005.03.078. [DOI] [PubMed] [Google Scholar]
- 115.Aziz MH, Hafeez BB, Sand JM, Pierce DB, Aziz SW, Dreckschmidt NE, Verma AK. Protein kinase Cvarepsilon mediates Stat3Ser727 phosphorylation, Stat3-regulated gene expression, and cell invasion in various human cancer cell lines through integration with MAPK cascade (RAF-1, MEK1/2, and ERK1/2) Oncogene. 2010;29:3100–3109. doi: 10.1038/onc.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cheng Y, LeGall T, Oldfield CJ, Mueller JP, Van YY, Romero P, Cortese MS, Uversky VN, Dunker AK. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 117.Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 118.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 119.Levine AJ, Hu W, Feng Z. The P53 pathway: what questions remain to be explored? Cell Death Differ. 2006;13:1027–1036. doi: 10.1038/sj.cdd.4401910. [DOI] [PubMed] [Google Scholar]
- 120.Yue X, Zhao Y, Xu Y, Zheng M, Feng Z, Hu W. Mutant p53 in Cancer: Accumulation, Gain-of-Function, and Therapy. J Mol Biol. 2017;429:1595–1606. doi: 10.1016/j.jmb.2017.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ozaki T, Nakagawara A. Role of p53 in Cell Death and Human Cancers. Cancers (Basel) 2011;3:994–1013. doi: 10.3390/cancers3010994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi PP, Will H, Schneider C, Del Sal G. Regulation of p53 activity in nuclear bodies by a specific PML isoform. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108–121. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Coltri PP, Dos Santos MGP, da Silva GHG. Splicing and cancer: Challenges and opportunities. Wiley Interdiscip Rev RNA. 2019;10:e1527. doi: 10.1002/wrna.1527. [DOI] [PubMed] [Google Scholar]
- 125.Dvinge H, Kim E, Abdel-Wahab O, Bradley RK. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16:413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Paronetto MP, Passacantilli I, Sette C. Alternative splicing and cell survival: from tissue homeostasis to disease. Cell Death Differ. 2016;23:1919–1929. doi: 10.1038/cdd.2016.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Di C, Syafrizayanti, Zhang Q, Chen Y, Wang Y, Zhang X, Liu Y, Sun C, Zhang H, Hoheisel JD. Function, clinical application, and strategies of Pre-mRNA splicing in cancer. Cell Death Differ. 2019;26:1181–1194. doi: 10.1038/s41418-018-0231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bressan GC, Moraes EC, Manfiolli AO, Kuniyoshi TM, Passos DO, Gomes MD, Kobarg J. Arginine methylation analysis of the splicing-associated SR protein SFRS9/SRP30C. Cell Mol Biol Lett. 2009;14:657–669. doi: 10.2478/s11658-009-0024-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Fu Y, Huang B, Shi Z, Han J, Wang Y, Huangfu J, Wu W. SRSF1 and SRSF9 RNA binding proteins promote Wnt signalling-mediated tumorigenesis by enhancing β-catenin biosynthesis. EMBO Mol Med. 2013;5:737–750. doi: 10.1002/emmm.201202218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lai CH, Huang YC, Lee JC, Tseng JT, Chang KC, Chen YJ, Ding NJ, Huang PH, Chang WC, Lin BW, Chen RY, Wang YC, Lai YC, Hung LY. Translational upregulation of Aurora-A by hnRNP Q1 contributes to cell proliferation and tumorigenesis in colorectal cancer. Cell Death Dis. 2017;8:e2555. doi: 10.1038/cddis.2016.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Perrotti D, Neviani P. From mRNA metabolism to cancer therapy: chronic myelogenous leukemia shows the way. Clin Cancer Res. 2007;13:1638–1642. doi: 10.1158/1078-0432.CCR-06-2320. [DOI] [PubMed] [Google Scholar]
- 132.Kiledjian M, Dreyfuss G. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 1992;11:2655–2664. doi: 10.1002/j.1460-2075.1992.tb05331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yoshino H, Enokida H, Chiyomaru T, Tatarano S, Hidaka H, Yamasaki T, Gotannda T, Tachiwada T, Nohata N, Yamane T, Seki N, Nakagawa M. Tumor suppressive microRNA-1 mediated novel apoptosis pathways through direct inhibition of splicing factor serine/arginine-rich 9 (SRSF9/SRp30c) in bladder cancer. Biochem Biophys Res Commun. 2012;417:588–593. doi: 10.1016/j.bbrc.2011.12.011. [DOI] [PubMed] [Google Scholar]
- 134.Han N, Li W, Zhang M. The function of the RNA-binding protein hnRNP in cancer metastasis. J Cancer Res Ther. 2013;9 Suppl:S129–S134. doi: 10.4103/0973-1482.122506. [DOI] [PubMed] [Google Scholar]
- 135.Cammas A, Pileur F, Bonnal S, Lewis SM, Lévêque N, Holcik M, Vagner S. Cytoplasmic relocalization of heterogeneous nuclear ribonucleoprotein A1 controls translation initiation of specific mRNAs. Mol Biol Cell. 2007;18:5048–5059. doi: 10.1091/mbc.E07-06-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Revil T, Pelletier J, Toutant J, Cloutier A, Chabot B. Heterogeneous nuclear ribonucleoprotein K represses the production of pro-apoptotic Bcl-xS splice isoform. J Biol Chem. 2009;284:21458–21467. doi: 10.1074/jbc.M109.019711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Xiao Z, Ko HL, Goh EH, Wang B, Ren EC. hnRNP K suppresses apoptosis independent of p53 status by maintaining high levels of endogenous caspase inhibitors. Carcinogenesis. 2013;34:1458–1467. doi: 10.1093/carcin/bgt085. [DOI] [PubMed] [Google Scholar]
- 138.Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L, Bramard A, Paquet ER, Watier V, Durand M, Lucier JF, Gervais-Bird J, Tremblay K, Prinos P, Klinck R, Elela SA, Chabot B. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol Cell Biol. 2008;28:6033–6043. doi: 10.1128/MCB.00726-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Haley B, Paunesku T, Protić M, Woloschak GE. Response of heterogeneous ribonuclear proteins (hnRNP) to ionising radiation and their involvement in DNA damage repair. Int J Radiat Biol. 2009;85:643–655. doi: 10.1080/09553000903009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Sueoka E, Sueoka N, Iwanaga K, Sato A, Suga K, Hayashi S, Nagasawa K, Nakachi K. Detection of plasma hnRNP B1 mRNA, a new cancer biomarker, in lung cancer patients by quantitative real-time polymerase chain reaction. Lung Cancer. 2005;48:77–83. doi: 10.1016/j.lungcan.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 141.Bouchal P, Dvorakova M, Scherl A, Garbis SD, Nenutil R, Vojtesek B. Intact protein profiling in breast cancer biomarker discovery: protein identification issue and the solutions based on 3D protein separation, bottom-up and top-down mass spectrometry. Proteomics. 2013;13:1053–1058. doi: 10.1002/pmic.201200121. [DOI] [PubMed] [Google Scholar]
- 142.Vu LP, Prieto C, Amin EM, Chhangawala S, Krivtsov A, Calvo-Vidal MN, Chou T, Chow A, Minuesa G, Park SM, Barlowe TS, Taggart J, Tivnan P, Deering RP, Chu LP, Kwon JA, Meydan C, Perales-Paton J, Arshi A, Gönen M, Famulare C, Patel M, Paietta E, Tallman MS, Lu Y, Glass J, Garret-Bakelman FE, Melnick A, Levine R, Al-Shahrour F, Järås M, Hacohen N, Hwang A, Garippa R, Lengner CJ, Armstrong SA, Cerchietti L, Cowley GS, Root D, Doench J, Leslie C, Ebert BL, Kharas MG. Functional screen of MSI2 interactors identifies an essential role for SYNCRIP in myeloid leukemia stem cells. Nat Genet. 2017;49:866–875. doi: 10.1038/ng.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Schaner ME, Ross DT, Ciaravino G, Sorlie T, Troyanskaya O, Diehn M, Wang YC, Duran GE, Sikic TL, Caldeira S, Skomedal H, Tu IP, Hernandez-Boussard T, Johnson SW, O'Dwyer PJ, Fero MJ, Kristensen GB, Borresen-Dale AL, Hastie T, Tibshirani R, van de Rijn M, Teng NN, Longacre TA, Botstein D, Brown PO, Sikic BI. Gene expression patterns in ovarian carcinomas. Mol Biol Cell. 2003;14:4376–4386. doi: 10.1091/mbc.E03-05-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Serce NB, Boesl A, Klaman I, von Serényi S, Noetzel E, Press MF, Dimmler A, Hartmann A, Sehouli J, Knuechel R, Beckmann MW, Fasching PA, Dahl E. Overexpression of SERBP1 (Plasminogen activator inhibitor 1 RNA binding protein) in human breast cancer is correlated with favourable prognosis. BMC Cancer. 2012;12:597. doi: 10.1186/1471-2407-12-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Guo K, Zheng S, Xu Y, Xu A, Chen B, Wen Y. Loss of miR-26a-5p promotes proliferation, migration, and invasion in prostate cancer through negatively regulating SERBP1. Tumour Biol. 2016;37:12843–12854. doi: 10.1007/s13277-016-5158-z. [DOI] [PubMed] [Google Scholar]
- 146.Hlavaty J, Ertl R, Miller I, Gabriel C. Expression of Progesterone Receptor Membrane Component 1 (PGRMC1), Progestin and AdipoQ Receptor 7 (PAQPR7), and Plasminogen Activator Inhibitor 1 RNA-Binding Protein (PAIRBP1) in Glioma Spheroids In Vitro. Biomed Res Int. 2016;2016:8065830. doi: 10.1155/2016/8065830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sun W, Guo C, Meng X, Yu Y, Jin Y, Tong D, Geng J, Huang Q, Qi J, Liu A, Guan R, Xu L, Sun D, Ji W, Liu P, Liu F, Sun H, Ji G, Fu S, Bai J. Differential expression of PAI-RBP1, C1orf142, and COTL1 in non-small cell lung cancer cell lines with different tumor metastatic potential. J Invest Med. 2012;60:689–694. doi: 10.2310/JIM.0b013e31824963b6. [DOI] [PubMed] [Google Scholar]
- 148.Lange CA, Sartorius CA, Abdel-Hafiz H, Spillman MA, Horwitz KB, Jacobsen BM. Progesterone receptor action: translating studies in breast cancer models to clinical insights. Adv Exp Med Biol. 2008;630:94–111. [PubMed] [Google Scholar]
- 149.Zhang L, Kanda Y, Roberts DJ, Ecker JL, Losel R, Wehling M, Peluso JJ, Pru JK. Expression of progesterone receptor membrane component 1 and its partner serpine 1 mRNA binding protein in uterine and placental tissues of the mouse and human. Mol Cell Endocrinol. 2008;287:81–89. doi: 10.1016/j.mce.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 150.Heaton JH, Dlakic WM, Gelehrter TD. Posttranscriptional regulation of PAI-1 gene expression. Thromb Haemost. 2003;89:959–966. [PubMed] [Google Scholar]
- 151.Harbeck N, Thomssen C. [u-Plasminogen activator (urinary plasminogen activator, urokinase) (uPA) and its PA-1 type 1 inhibitor are not only prognostically but also predictively significant and support clinical decisions on therapy in primary carcinoma of the breast] Zentralbl Gynakol. 2003;125:362–367. doi: 10.1055/s-2003-43036. [DOI] [PubMed] [Google Scholar]
- 152.Kuhn W, Schmalfeldt B, Reuning U, Pache L, Berger U, Ulm K, Harbeck N, Späthe K, Dettmar P, Höfler H, Jänicke F, Schmitt M, Graeff H. Prognostic significance of urokinase (uPA) and its inhibitor PAI-1 for survival in advanced ovarian carcinoma stage FIGO IIIc. Br J Cancer. 1999;79:1746–1751. doi: 10.1038/sj.bjc.6690278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tecimer C, Doering DL, Goldsmith LJ, Meyer JS, Abdulhay G, Wittliff JL. Clinical relevance of urokinase-type plasminogen activator, its receptor and inhibitor type 1 in ovarian cancer. Int J Gynecol Cancer. 2000;10:372–381. doi: 10.1046/j.1525-1438.2000.010005372.x. [DOI] [PubMed] [Google Scholar]
- 154.Schmalfeldt B, Prechtel D, Härting K, Späthe K, Rutke S, Konik E, Fridman R, Berger U, Schmitt M, Kuhn W, Lengyel E. Increased expression of matrix metalloproteinases (MMP)-2, MMP-9, and the urokinase-type plasminogen activator is associated with progression from benign to advanced ovarian cancer. Clin Cancer Res. 2001;7:2396–2404. [PubMed] [Google Scholar]
- 155.Borgfeldt C, Hansson SR, Gustavsson B, Måsbäck A, Casslén B. Dedifferentiation of serous ovarian cancer from cystic to solid tumors is associated with increased expression of mRNA for urokinase plasminogen activator (uPA), its receptor (uPAR) and its inhibitor (PAI-1) Int J Cancer. 2001;92:497–502. doi: 10.1002/ijc.1215. [DOI] [PubMed] [Google Scholar]
- 156.Hoffmann G, Pollow K, Weikel W, Strittmatter HJ, Bach J, Schaffrath M, Knapstein P, Melchert F, Pollow B. Urokinase and plasminogen activator-inhibitor (PAI-1) status in primary ovarian carcinomas and ovarian metastases compared to benign ovarian tumors as a function of histopathological parameters. Clin Chem Lab Med. 1999;37:47–54. doi: 10.1515/CCLM.1999.007. [DOI] [PubMed] [Google Scholar]
- 157.van der Burg ME, Henzen-Logmans SC, Berns EM, van Putten WL, Klijn JG, Foekens JA. Expression of urokinase-type plasminogen activator (uPA) and its inhibitor PAI-1 in benign, borderline, malignant primary and metastatic ovarian tumors. Int J Cancer. 1996;69:475–479. doi: 10.1002/(SICI)1097-0215(19961220)69:6<475::AID-IJC10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- 158.Henderson MJ, Russell AJ, Hird S, Muñoz M, Clancy JL, Lehrbach GM, Calanni ST, Jans DA, Sutherland RL, Watts CK. EDD, the human hyperplastic discs protein, has a role in progesterone receptor coactivation and potential involvement in DNA damage response. J Biol Chem. 2002;277:26468–26478. doi: 10.1074/jbc.M203527200. [DOI] [PubMed] [Google Scholar]
- 159.Fogli A, Gauthier-Barichard F, Schiffmann R, Vanderhoof VH, Bakalov VK, Nelson LM, Boespflug-Tanguy O. Screening for known mutations in EIF2B genes in a large panel of patients with premature ovarian failure. BMC Womens Health. 2004;4:8. doi: 10.1186/1472-6874-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Choi Y, Rosewell KL, Brännström M, Akin JW, Curry TE, Jr, Jo M. FOS, a Critical Downstream Mediator of PGR and EGF Signaling Necessary for Ovulatory Prostaglandins in the Human Ovary. J Clin Endocrinol Metab. 2018;103:4241–4252. doi: 10.1210/jc.2017-02532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Mansouri MR, Schuster J, Badhai J, Stattin EL, Lösel R, Wehling M, Carlsson B, Hovatta O, Karlström PO, Golovleva I, Toniolo D, Bione S, Peluso J, Dahl N. Alterations in the expression, structure and function of progesterone receptor membrane component-1 (PGRMC1) in premature ovarian failure. Hum Mol Genet. 2008;17:3776–3783. doi: 10.1093/hmg/ddn274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Peluso JJ, Griffin D, Liu X, Horne M. Progesterone receptor membrane component-1 (PGRMC1) and PGRMC-2 interact to suppress entry into the cell cycle in spontaneously immortalized rat granulosa cells. Biol Reprod. 2014;91:104. doi: 10.1095/biolreprod.114.122986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yilmaz BD, Bulun SE. Endometriosis and nuclear receptors. Hum Reprod Update. 2019;25:473–485. doi: 10.1093/humupd/dmz005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Alkhalaf M, El-Mowafy AM, Abou-Zeid LA. Progesterone inhibition of MDM2 p90 protein in MCF-7 human breast cancer cell line is dependent on p53 levels. J Mol Genet Med. 2005;1:33–37. doi: 10.4172/1747-0862.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Chen Y, Wu F, Saito E, Lin Y, Song M, Luu HN, Gupta PC, Sawada N, Tamakoshi A, Shu XO, Koh WP, Xiang YB, Tomata Y, Sugiyama K, Park SK, Matsuo K, Nagata C, Sugawara Y, Qiao YL, You SL, Wang R, Shin MH, Pan WH, Pednekar MS, Tsugane S, Cai H, Yuan JM, Gao YT, Tsuji I, Kanemura S, Ito H, Wada K, Ahn YO, Yoo KY, Ahsan H, Chia KS, Boffetta P, Zheng W, Inoue M, Kang D, Potter JD. Association between type 2 diabetes and risk of cancer mortality: a pooled analysis of over 771,000 individuals in the Asia Cohort Consortium. Diabetologia. 2017;60:1022–1032. doi: 10.1007/s00125-017-4229-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Michlig S, Harris M, Loffing J, Rossier BC, Firsov D. Progesterone down-regulates the open probability of the amiloride-sensitive epithelial sodium channel via a Nedd4-2-dependent mechanism. J Biol Chem. 2005;280:38264–38270. doi: 10.1074/jbc.M506308200. [DOI] [PubMed] [Google Scholar]
- 167.Boyan BD, Schwartz Z. Rapid vitamin D-dependent PKC signaling shares features with estrogen-dependent PKC signaling in cartilage and bone. Steroids. 2004;69:591–597. doi: 10.1016/j.steroids.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 168.Wickramasekera NT, Das GM. Tumor suppressor p53 and estrogen receptors in nuclear-mitochondrial communication. Mitochondrion. 2014;16:26–37. doi: 10.1016/j.mito.2013.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]