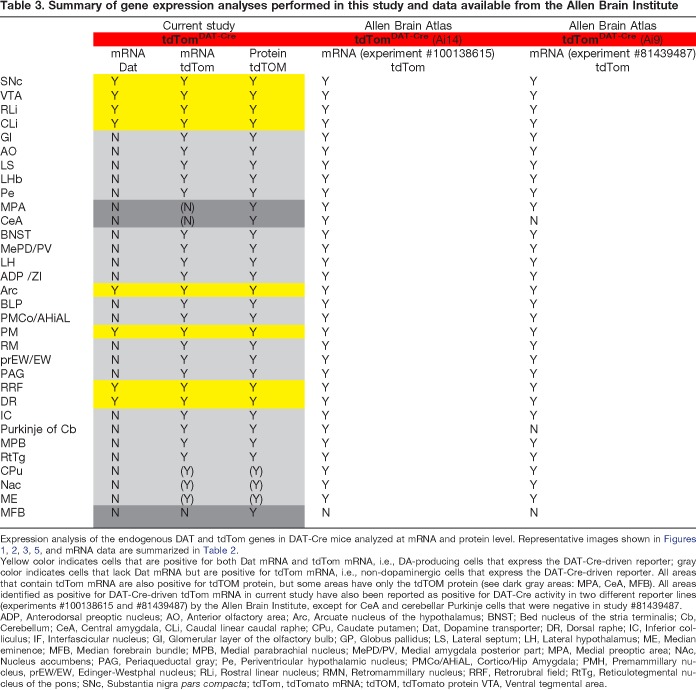
Table 3.
Summary of gene expression analyses performed in this study and data available from the Allen Brain Institute
Expression analysis of the endogenous DAT and tdTom genes in DAT-Cre mice analyzed at mRNA and protein level. Representative images shown in Figures 1, 2, 3, 5, and mRNA data are summarized in Table 2.
Yellow color indicates cells that are positive for both Dat mRNA and tdTom mRNA, i.e., DA-producing cells that express the DAT-Cre-driven reporter; gray color indicates cells that lack Dat mRNA but are positive for tdTom mRNA, i.e., non-dopaminergic cells that express the DAT-Cre-driven reporter. All areas that contain tdTom mRNA are also positive for tdTOM protein, but some areas have only the tdTOM protein (see dark gray areas: MPA, CeA, MFB). All areas identified as positive for DAT-Cre-driven tdTom mRNA in current study have also been reported as positive for DAT-Cre activity in two different reporter lines (experiments #100138615 and #81439487) by the Allen Brain Institute, except for CeA and cerebellar Purkinje cells that were negative in study #81439487.
ADP, Anterodorsal preoptic nucleus; AO, Anterior olfactory area; Arc, Arcuate nucleus of the hypothalamus; BNST; Bed nucleus of the stria terminalis; Cb, Cerebellum; CeA, Central amygdala, CLi, Caudal linear caudal raphe; CPu, Caudate putamen; Dat, Dopamine transporter; DR, Dorsal raphe; IC, Inferior colliculus; IF, Interfascicular nucleus; GI, Glomerular layer of the olfactory bulb; GP, Globus pallidus; LS, Lateral septum; LH, Lateral hypothalamus; ME, Median eminence; MFB, Median forebrain bundle; MPB, Medial parabrachial nucleus; MePD/PV, Medial amygdala posterior part; MPA, Medial preoptic area; NAc, Nucleus accumbens; PAG, Periaqueductal gray; Pe, Periventricular hypothalamic nucleus; PMCo/AHiAL, Cortico/Hip Amygdala; PMH, Premammillary nucleus, prEW/EW, Edinger-Westphal nucleus; RLi, Rostral linear nucleus; RMN, Retromammillary nucleus; RRF, Retrorubral field; RtTg, Reticulotegmental nucleus of the pons; SNc, Substantia nigra pars compacta; tdTom, tdTomato mRNA; tdTOM, tdTomato protein VTA, Ventral tegmental area.